- Title
-
Evolution of two-pore domain potassium channels and their gene expression in zebrafish embryos
- Authors
- Park, S.J., Silic, M.R., Staab, P.L., Chen, J., Zackschewski, E.L., Zhang, G.
- Source
- Full text @ Dev. Dyn.
|
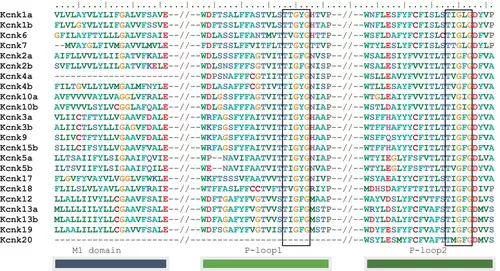
Alignment of zebrafish key two-pore domain potassium (K2P) channel domains. zebrafish K2p channels possess two typical pore-forming loops (P-loops). Only sequences around these domains were shown. The double dashes indicate the noncontinued sequences. M1 is the first transmembrane domain. Potassium filter domains (T-X-G-Y/F-G) were boxed. Amino acids, L and I, are presented in the filter domain of the second loop. The short Kcnk4a and Kcnk20 sequences may be caused by incomplete genome annotation of GRCz11 (Genome Refrence Consortium zebrafish reference genome 11). |
|
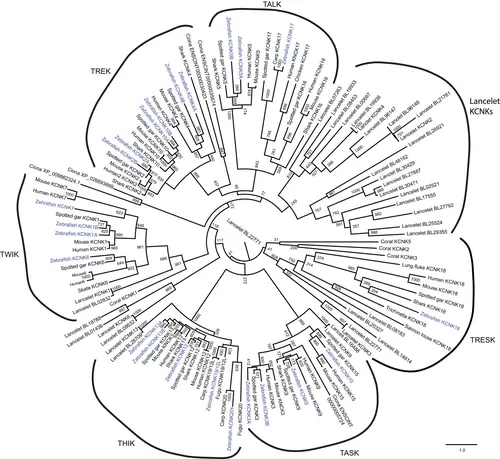
Maximum likelihood phylogeny of two-pore domain potassium (K2P) channels. Some representative species (human, mouse, zebrafish, spotted gar, shark, lancelet, tunicate, and coral) were analyzed. Numbers at each node indicate the supporting values based on 1000 bootstrap replicates. Branch lengths are proportional to expected replacements per site. The phylogeny was inferred with the Jones–Taylor–Thornton model model plus gamma distribution using PhyML. All the six subtypes of K2P channels were grouped together and indicated with curved lines. The zebrafish K2p channels were highlighted in blue. Many lancelet K2P channels formed a distinct group on the phylogenetic tree. TALK, TWIK-related alkaline pH-activated K+; TASK, TWIK-related acid-sensitive K+; THIK, tandem pore domain halothane-inhibited K+; TREK, TWIK-related K+; TRESK, TWIK-related spinal cord K+; TWIK, tandem of pore domains in a weak inward rectifying K+. |
|
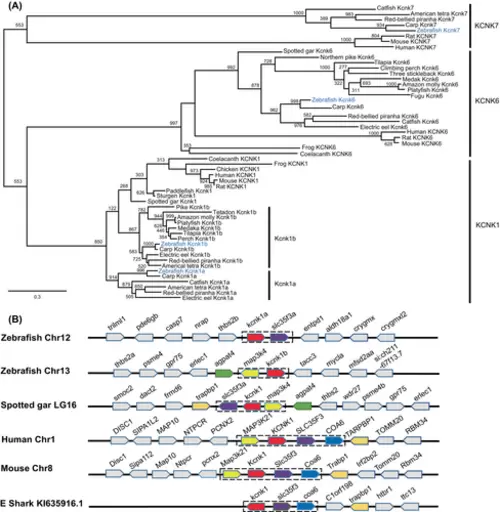
The kcnk1 gene was duplicated in teleosts, including zebrafish. (A) Molecular phylogenetic tree generated using maximum likelihood analysis on vertebrate zebrafish tandem of pore domains in a weak inward rectifying K+ (TWIK) channel proteins as obtained with Jones–Taylor–Thornton model plus gamma distribution. Numbers at each node denote bootstrap values based on 1000 replicates. Branch lengths are proportional to expected replacements per site. All the zebrafish TWIK channels are highlighted in blue. KCNK1, KCNK6, and KCNK7 formed distinct clades. (B) The synteny of KCNK1 in five representative vertebrate species. The illustration of the genes and their sizes are not proportional to the length of the distances between genes. KCNK1 was highlighted in red. The conserved synteny (map3k4–kcnk1–slc35f3–coa6) was boxed with dashed lines. |
|
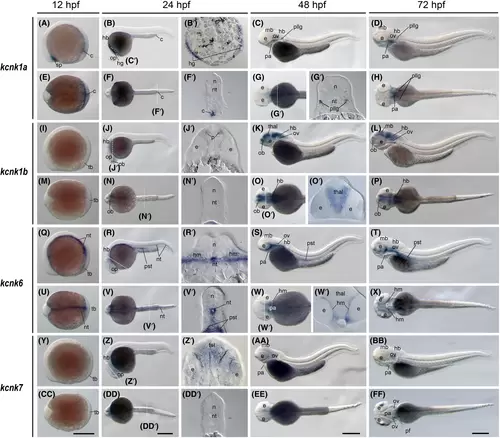
The kcnk1a, kcnk1b, kcnk6, and kcnk7 gene (tandem of pore domains in a weak inward rectifying K+ [TWIK] channel) expression during zebrafish embryogenesis. Whole-mount in situ hybridization of zebrafish embryos at stages 12 h postfertilization (hpf) (A, E, I, M, Q, U, Y, and CC), 24 hpf (B, F, J, N, R, V, Z, and DD), 48 hpf (C, G, K, O, S, W, AA, EE), and 72 hpf (D, H, L, P, T, X, BB, and FF). The anterior is to the left in all the whole-mount images, and the dorsal is to the top in all transverse sections (B′, F′, G′, J′, N′, O′, R′, V′, W′, Z', and DD'). (A–D) Lateral view of gene expression of kcnk1a. (E–H) Dorsal view of gene expression of kcnk1a. (I–L) Lateral view of gene expression of kcnk1b. (M–P) Dorsal view of gene expression of kcnk1b. (Q–T) Lateral view of gene expression of kcnk6. (U–X) Dorsal view of gene expression of kcnk6. (Y–BB) Lateral view of gene expression of kcnk7. (CC–FF) Dorsal view of gene expression of kcnk7. The white dashed lines indicate the positions of sections left to the panel. The letters below or around the dashed lines correspond to the section panels. Scale bars were added on the bottom row of images; 250 μm for whole-mount images. c, cloaca; e, eye; hb, hindbrain; hg, hatching gland; hm, head mesenchymal region; mb, midbrain; n, neural tube; nt, notochord; ob, olfactory bulbs; op, optic vesicles; ov, otic vesicles; pa, pharyngeal arches; pf, pectoral fins; pllg, posterior lateral line ganglia; pst, pronephric proximal straight tubule; tb, tail bud; tel, telencephalon; thal, thalamus. |
|
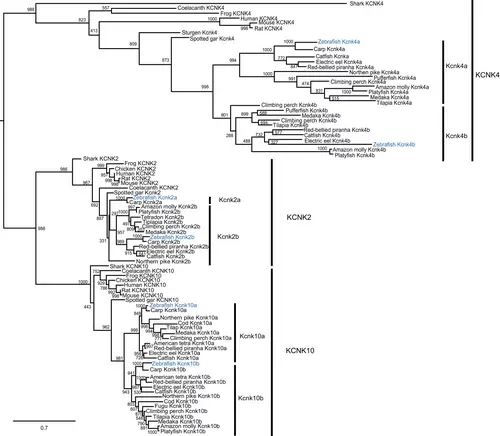
The kcnk2, kcnk4, and kcnk10 genes were duplicated in teleosts, including zebrafish. The molecular phylogenetic tree was generated using maximum likelihood analysis on vertebrate tandem of pore domains in a weak inward rectifying K+ channel-related K+ (TREK) channel proteins as obtained with Jones–Taylor–Thornton model plus gamma distribution. Numbers at each node denote bootstrap values based on 1000 replicates. Branch lengths are proportional to expected replacements per site. All the TREK members, KCNK2, KCNK4, and KCNK10 formed distinct clades. Teleost channel formed two subgroups among each TREK channel su-bclade. The zebrafish channels were highlighted in blue. |
|
The syntenic analyses on the duplicated kcnk2 and kcnk10 genes in five representative vertebrate species. The illustration of the genes and their sizes are not proportional to the length of the distances between genes. (A) Common vertebrate kcnk2 synteny. KCNK2 was highlighted in red. The synteny (kcnk2–cenfp) is shared among sharks, mice, and humans. In comparison, the zebrafish and spotted gar share the kcnk2–cgref1–prkg3 synteny. The zebrafish kcnk2a is still linked with cenpf, like sharks and humans. (B) Common vertebrate kcnk10 synteny. KCNK10, is highlighted in red. The synteny (kcnk10–gpr65) was boxed with blue dashed lines. This synteny presents in all five species. The spotted gar shares a longer synteny with zebrafish. |
|
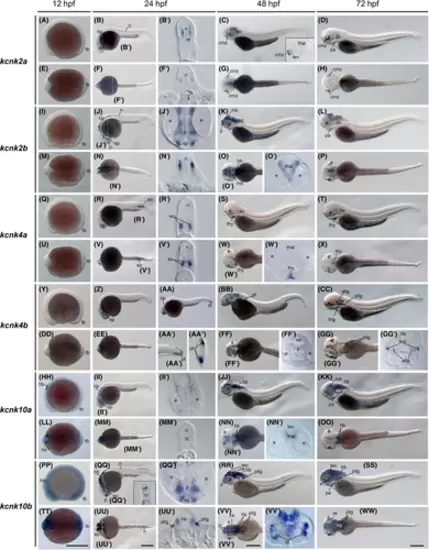
The kcnk2a, kcnk2b, kcnk4a, kcnk4b, kcnk10a, and kcnk10b gene (tandem of pore domains in a weak inward rectifying K+-related K+ channel [TREK]) expression during zebrafish embryogenesis. Whole-mount in situ hybridization of zebrafish embryos at stages 12 h postfertilization (hpf) (A, E, I, M, Q, U, Y, DD, HH, LL, PP, and TT), 24 hpf (B, F, J, N, R, V, Z, EE, II, MM, QQ, and UU), 36 hpf (AA–AA”); 48 hpf (C, G, K, O, S, W, BB, FF, JJ, NN, RR, and VV), and 72 hpf (D, H, L, P, T, X, CC, GG, KK, OO, SS, and WW). The anterior is to the left in all the whole-mount images, and the dorsal is to the top in all transverse sections (B′, F′, J′, N′, O′, R′, V′, W′, AA′′, FF′, GG′, II′, MM', NN', QQ', UU', V and V'). (A–D) Lateral view of gene expression of kcnk2a. (E–H) Dorsal view of gene expression of kcnk2a. (I–L) Lateral view of gene expression of kcnk2b. (M–P) Dorsal view of gene expression of kcnk2b. (Q–T) Lateral view of gene expression of kcnk4a. (U–X) Dorsal view of gene expression of kcnk4a. (Y–CC) Lateral view of gene expression of kcnk4b. (DD–GG) Dorsal view of gene expression of kcnk4b. (HH–KK) Lateral view of gene expression of kcnk10a. (LL–OO) Dorsal view of gene expression of kcnk10a. (PP–SS) Lateral view of gene expression of kcnk10b. (TT–WW) Dorsal view of gene expression of kcnk10b. The white dashed lines indicate the positions of sections. The letters below or around the dashed lines correspond to the section panels. Scale bars were added on the bottom row of images; 250 μm for whole-mount images. allg, anterior lateral line ganglia; ce, cerebellum; cf, caudal fin fold; cmz, ciliary marginal zones; e, eye; hb, hindbrain; mb, midbrain; n, neural tube; nt, notochord; op, optic vesicles; ov, otic vesicles; p, pallium; pa, pharyngeal arches; pf, pectoral fin; pllg, posterior lateral line ganglia; re, retina; so, somites; sp, subpallium; tb, tail bud; tec, tectum; thal, thalamus; thy, thyroid gland; trig, trigeminal ganglia. |
|
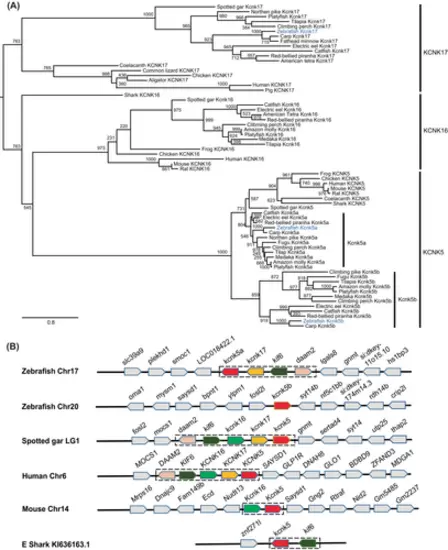
The kcnk5 gene was duplicated in teleosts, including zebrafish. (A) Molecular phylogenetic tree generated using maximum likelihood analysis on vertebrate tandem of pore domains in a weak inward rectifying K+-related alkaline pH-activated K+ (TALK) channel proteins as obtained with Jones–Taylor–Thornton model plus gamma distribution. Numbers at each node denote bootstrap values based on 1000 replicates. Branch lengths are proportional to expected replacements per site. The zebrafish channels were highlighted in blue. The TALK channels, KCNK5, KCNK16, and KCNK17 formed distinct clades. The Kcnk5a and Kcnk5b formed two subgroups within the KCNK5 clade. (B) The synteny of KCNK5 in five representative vertebrate species. The illustration of the genes and their sizes are not proportional to the length of the distances between genes. KCNK5 was highlighted in red. The conserved synteny (daam2–kif6–kcnk16–kcnk17–kcnk5) was boxed with dashed lines. This synteny in elephant shark is short due to limited available genomic information. |
|
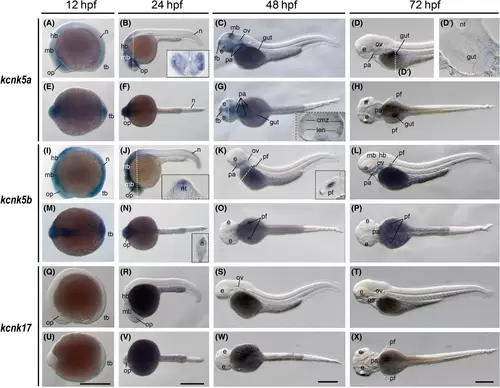
The kcnk5a, kcnk5b, and kcnk17 gene (tandem of pore domains in a weak inward rectifying K+-related alkaline pH-activated K+ channel) expression during zebrafish embryogenesis. Whole-mount in situ hybridization of zebrafish embryos at stages 12 h postfertilization (hpf) (A, E, I, M, Q, and U), 24 hpf (B, F, J, N, R, and V), 48 hpf (C, G, K, O, S, and W), and 72 hpf (D, H, L, P, T, and X). The anterior is to the left in all the whole-mount images, and the dorsal is to the top in all transverse sections. The section images were shown in the insert of corresponding panels (B, G, J, K, and N), or on the left side (D′). (A–D) Lateral view of gene expression of kcnk5a. The gut is labeled with orange dashed lines in panel D′. (E–H) Dorsal view of gene expression of kcnk5a. (I–L) Lateral view of gene expression of kcnk5b. (M–P) Dorsal view of gene expression of kcnk5b. (Q–T) Lateral view of gene expression of kcnk17. (U–X) Dorsal view of gene expression of kcnk17. The white dashed lines indicate the positions of sections. The section images were shown in the insert of corresponding panels. Scale bars were added on the bottom row of images; 250 μm for whole-mount images. cmz, ciliary marginal zones; e, eye; fb, forebrain; gut, gut; hb, hindbrain; len, lens; mb, midbrain; n, neural tube; nt, notochord; op, optic vesicles; ov, otic vesicles; pa, pharyngeal arches; pf, pectoral fins; tb, tail bud. |
|
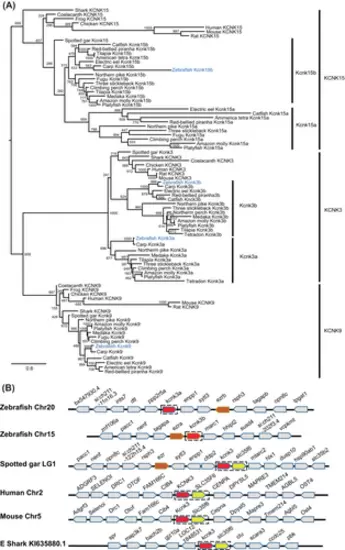
The kcnk3 and kcnk15 genes were duplicated in teleosts, including zebrafish. (A) Molecular phylogenetic tree generated using maximum likelihood analysis on vertebrate tandem of pore domains in a weak inward rectifying K+-related acid-sensitive K+ (TASK) channel proteins as obtained with Jones–Taylor–Thornton model plus gamma distribution. Numbers at each node denote bootstrap values based on 1000 replicates. Branch lengths are proportional to expected replacements per site. The KCNK3, KCNK9, and KCNK13 channels formed distinct clades. The zebrafish TASK channels were highlighted in blue. Teleost Kcnk3 and Kcnk15 channels formed two subbranches within the KCNK3 and KCNK15 clade, respectively. (B) The synteny of KCNK3 in five representative vertebrate species. The illustration of the genes and their sizes are not proportional to the length of the distances between genes. KCNK3 was highlighted in red. The conserved synteny (kcnk3–slc35f6) was boxed with dashed lines. |
|
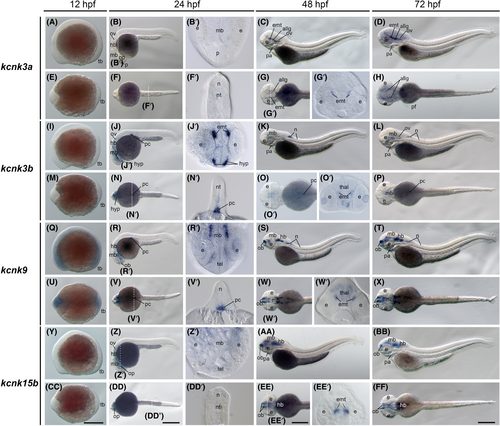
The kcnk3a, kcnk3b, kcnk9, and kcnk15b gene (tandem of pore domains in a weak inward rectifying K+-related acid-sensitive K+ [TASK] channel) expression during zebrafish embryogenesis. Whole-mount in situ hybridization of zebrafish embryos at stages 12 h postfertilization (hpf) (A, E, I, M, Q, U, Y, and CC), 24 hpf (B, F, J, N, R, V, Z, and DD), 48 hpf (C, G, K, O, S, W, AA, and EE), and 72 hpf (D, H, L, P, T, X, BB, and FF). The anterior is to the left in all the whole-mount images, and the dorsal is to the top in all transverse sections (B′, F′, G2, J′, N′, O′, R′, V′, W′, Z′, DD′, and EE′). (A–D) Lateral view of gene expression of kcnk3a. (E–H) Dorsal view of gene expression of kcnk3a. (I–L) Lateral view of gene expression of kcnk3b. (M–P) Dorsal view of gene expression of kcnk3b. (Q–T) Lateral view of gene expression of kcnk9. (U–X) Dorsal view of gene expression of kcnk9. (Y–BB) Lateral view of gene expression of kcnk15b. (CC–FF) Dorsal view of gene expression of kcnk15b. The white dashed lines indicate the positions of sections. The letters below or around the dashed lines correspond to the panels. Scale bars were added on the bottom row of images; 250 μm for whole-mount images. allg, anterior lateral line ganglia; e, eye; emt, eminentia thalami; hb, hindbrain; hyp, hypothalamus; mb, midbrain; n, neural tube; nt, notochord; ob, olfactory bulbs; op, optic vesicles; ov, otic vesicles; p, pallium; pa, pharyngeal arches; pc, pancreas; pc, pancreas; pf, pectoral fins; tb, tail bud, tel, telencephalon; thal, thalamus. |
|
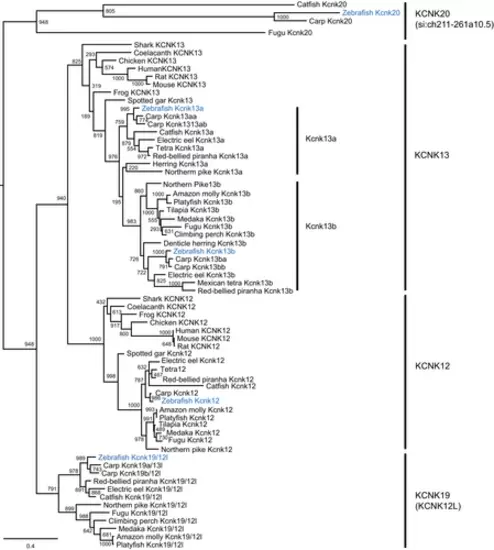
The kcnk13 gene was duplicated in teleosts, including zebrafish. The molecular phylogenetic tree was generated using maximum likelihood analysis on vertebrate tandem pore domain halothane-inhibited K+ (THIK) channel proteins with Jones–Taylor–Thornton model plus gamma distribution. Numbers at each node denote bootstrap values based on 1000 replicates. Branch lengths are proportional to expected replacements per site. All the THIK members, KCNK12, KCNK13, KNCK19, and KCNK20, formed distinct clades. Teleost channel formed two subgroups, Kcnk13a and Kcnk13b, within the KCNK13 tandem of pore domains in a weak inward rectifying K+-related K channel clade. The zebrafish channels were highlighted in blue. |
|
The syntenic analyses on the duplicated kcnk13, kcnk19, and kcnk20 genes in five representative vertebrate species. The illustration of the genes and their sizes are not proportional to the length of the distances between genes. (A) Common vertebrate kcnk13 synteny. KCNK13 was highlighted in red. The synteny (foxn3–efcab11–tdp1–kcnk13–pmsc1) is shared among the five species. (B) Shared kcnk19 synteny between zebrafish and common carp. (C) Shared kcnk20 synteny between zebrafish and fugu. The syntenies were boxed with blue dashed lines. |
|
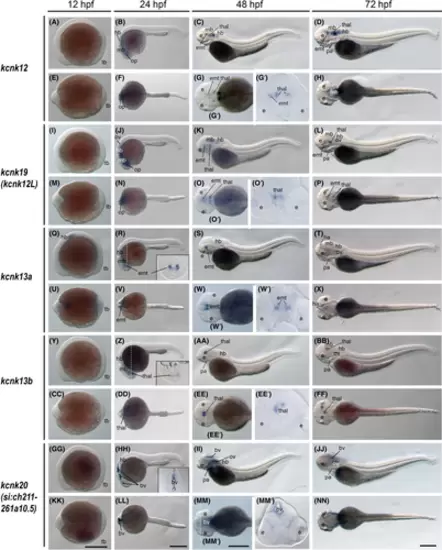
The kcnk12, kcnk19, kcnk13a, kcnk13b, and kcnk20 gene (tandem pore domain halothane-inhibited K+ channel) expression during zebrafish embryogenesis. Whole-mount in situ hybridization of zebrafish embryos at stages 12 h postfertilization (hpf) (A, E, I, M, Q, U, Y, CC, GG, and KK), 24 hpf (B, F, J, N, R, V, Z, DD, HH, and LL), 48 hpf (C, G, K, O, S, W, AA, EE, II, and MM), and 72 hpf (D, H, L, P, T, X, BB, FF, JJ, and NN). The anterior is to the left in all the whole-mount images, and the dorsal is to the top in all transverse sections. (A–D) Lateral view of gene expression of kcnk12. (E–H) Dorsal view of gene expression of kcnk12. (I–L) Lateral view of gene expression of kcnk19. (M–P) Dorsal view of gene expression of kcnk19. (Q–T) Lateral view of gene expression of kcnk13a. (U–X) Dorsal view of gene expression of kcnk13a. (Y–BB) Lateral view of gene expression of kcnk13b. (CC–FF) Dorsal view of gene expression of kcnk13b. (GG–JJ) Lateral view of gene expression of kcnk20. (KK–NN) Dorsal view of gene expression of kcnk20. The white dashed lines indicate the positions of sections. The letters below or around the dashed lines correspond to the panels. Scale bars were added on the bottom row of images; 250 μm for whole-mount images. bv, brain ventricles; e, eye; emt, eminentia thalami; ha, habenula; hb, hindbrain; mb, midbrain; op, optic vesicles; ov, otic vesicles; p, pallium; pa, pharyngeal arches; tb, tail bud; thal, thalamus. |
|
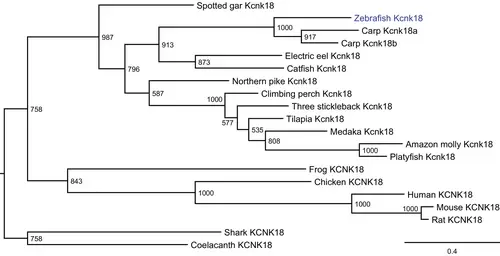
The kcnk18 gene is present in teleosts, including zebrafish. The molecular phylogenetic tree was generated using maximum likelihood analysis on vertebrate KCNK18 proteins as obtained with Jones–Taylor–Thornton model plus gamma distribution. Numbers at each node denote bootstrap values based on 1000 replicates. Branch lengths are proportional to expected replacements per site. The zebrafish Kcnk18 channel was highlighted in blue. |
|
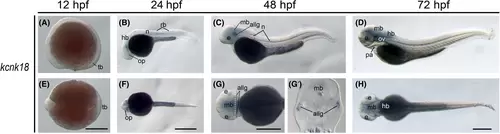
The kcnk18 gene (tandem of pore domains in a weak inward rectifying K+-related spinal cord K+ channel) expression during zebrafish embryogenesis. Whole-mount in situ hybridization of zebrafish embryos at stages 12 h postfertilization (hpf) (A and E), 24 hpf (B and F), 48 hpf (C and G), and 72 hpf (D and H). The anterior is to the left in all the whole-mount images, and the dorsal is to the top in all transverse sections (G2). (A–D) Lateral view of gene expression of kcnk18. (E–H) Dorsal view of gene expression of kcnk18. The white dashed lines indicate the positions of sections. The section images were shown in the insert of corresponding panels. Scale bars were added on the bottom row of images; 250 μm for whole-mount images. allg, anterior lateral line ganglia; e, eye; hb, hindbrain; mb, midbrain; n, neural tube; op, optic vesicles; ov, otic vesicles; pa, pharyngeal arches; rb, Rohon–Beard (RB) neurons; tb, tail bud. |
|
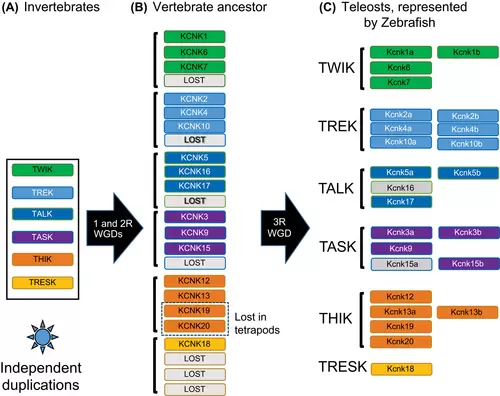
Model of vertebrate two-pore domain potassium (K2P) gene evolution. Based on our phylogenetic and syntenic analysis, we proposed this model for this two-pore potassium channel. (A) In invertebrates, the six subtypes of K2P have already evolved. At least there is one gene for each subtype. Some species, such as lancelets and tunicates, may get multiple copies of K2P genes through independent duplication events. (B) Two consecutive whole-genome duplications (WGDs) happened at the dawn of vertebrates. Then, all the K2P genes were doubled twice in the vertebrate ancestor. Some of the genes were lost after the two rounds of WGDs (2R). Lost genes are in gray boxes. It is worth noting that the KCNK19 and KCNK20 were lost in tetrapods. (C) The 3rd round teleost-specific WGD (3R) further reshaped the gene numbers in this group of animals. Some gene duplicates were maintained, whereas others were lost. In zebrafish, the kcnk15a and kcnk16 were lost independently (gray-colored boxes) but retained in other teleost fish. The horizontal black arrows indicate the WGD events. TALK, TWIK-related alkaline pH-activated K+; TASK, TWIK-related acid-sensitive K+; THIK, tandem pore domain halothane-inhibited K+; TREK, TWIK-related K+; TRESK, TWIK-related spinal cord K+; TWIK, tandem of pore domains in a weak inward rectifying K+. |
|
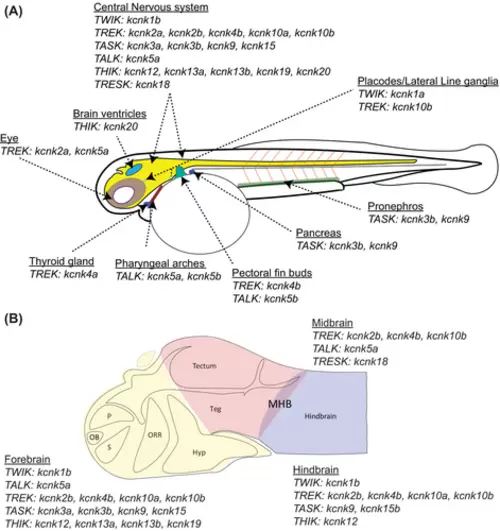
Schematic summary of two-pore domain potassium (k2p) gene expression in 48 h postfertilization (hpf) zebrafish embryos. (A) The zebrafish k2p gene expression is summarized for their expression in major organs or tissues. (B) The summary of zebrafish k2p genes that were detected in the brain. The forebrain, midbrain, and hindbrain are color-coded. The anatomic structures are made according to the atlas of embryonic zebrafish brain.51 Hyp, hypothalamus; MHB, midbrain–hindbrain boundary; OB, olfactory bulb, ORR, optic recess region; p, pallium; s, subpallium; TALK, TWIK-related alkaline pH-activated K+; TASK, TWIK-related acid-sensitive K+; Teg, tegmentum; THIK, tandem pore domain halothane-inhibited K+; TREK, TWIK-related K+; TRESK, TWIK-related spinal cord K+; TWIK, tandem of pore domains in a weak inward rectifying K+. |