- Title
-
Determining Photoreceptor Cell Identity: Rod Versus Cone Fate Governed by tbx2b Opposing nrl
- Authors
- Neil, G.J., Kluttig, K.H., Allison, W.T.
- Source
- Full text @ Invest. Ophthalmol. Vis. Sci.
|
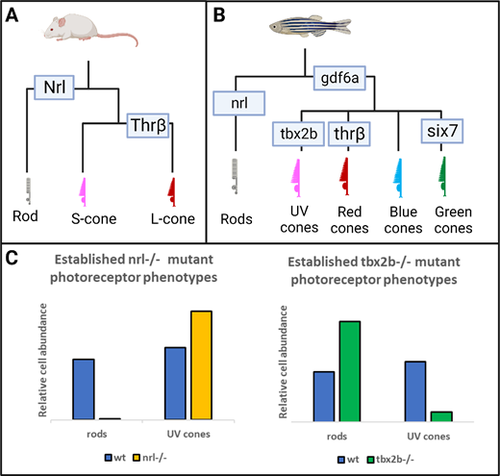
The nrl and tbx2b mutants have strikingly opposite effects on rod and sws1 cone developmental phenotypes. (A, B) Schematics highlighting that the role of Nrl in patterning the rod fate in mouse models is recapitulated and expanded upon in the zebrafish model. (C) Previous research has established that nrl mutants exhibit a dramatic loss of rod cell development and a concomitant increase of short-wavelength cones (mice and zebrafish),13,15 whereas the opposite is observed in tbx2b mutants (zebrafish).25 We seek to resolve whether this relation between the rod and UV cone fates represents a single fate determination mechanism, or two independent mechanisms governing each cell fate. |
|
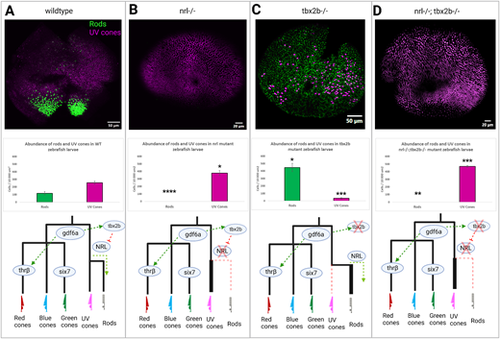
The nrl and tbx2b share an epistatic relationship governing rod versus UV cone fate. Schematic of putative interaction of tbx2b repressing activation of nrl in otherwise UV cone fated cells. (A) In wildtype zebrafish, nrl is sufficient to induce the rod fate but is repressed by tbx2b in UV fated cells. (B) Loss of nrl expression leads to failure to develop rods and increase in UV cone production (N = 4), as cells fail to adopt the rod fate and instead take on a default UV cone program. (C) Loss of tbx2b releases nrl from repression, driving an excess of rod development in otherwise UV cone fated cells (N = 3). (D) Double mutants recapitulate the single nrl mutant phenotype, producing excess UV cones at the expense of rods (N = 5) as tbx2b is not needed to act on an already non-functional nrl. Together these results indicate that tbx2b is not required for UV cone development, but instead likely prevents nrl expression from imposing the rod fate on UV fated cells. Rods and UV cones detected with antibodies 4C12 and 10C9.1, respectively, label an unknown rod epitope and Sws1 opsin. * = P < 0.05, ** = P < 0.005, *** = P < 0.0005 significance as determined by unpaired t-test. Significance markers denote differences in abundance of each photoreceptor cell type relative to wildtype. |
|
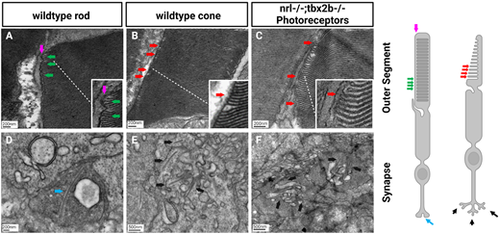
Electron microscopy reveals lack of rod-like outer segments or synapses in nrl−/−; tbx2b−/− double mutants. Using the metrics described below, no nrl−/−;tbx2b−/− double mutant larvae was found to have a cell that could confidently be described as a rod, despite assessing six individual larvae. In wild types, all six larvae examined had obvious rod photoreceptors. This reaffirms that although mutation of tbx2b promotes a rod-like phenotype, it is insufficient to rescue loss of nrl. Rod cells can be identified morphologically using EM from key distinctive features. Rod outer segments (A) are fully enclosed by the outer segment membrane (magenta arrow) and contain discrete stacked disks which can be identified by characteristic hairpin loops at their ends (green arrows). Cone outer segments (B, C) result from invaginations of the outer segment membrane, producing disk layers made of continuous membrane that are exposed to the extracellular matrix (red arrows). Rod synapses (D) are electron dense structures, appearing darker than the surrounding cone synapses, and contain a single, relatively large, synaptic ribbon (blue arrow). Cone synapses (E, F) are identifiable by an abundance of comparatively shorter synaptic ribbons (black arrows). PHENOTYPE:
|
|
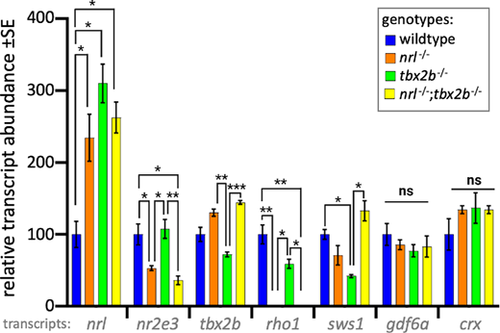
Photoreceptor transcripts in larval zebrafish show that nrl−/−; tbx2b−/− mutants recapitulate phenotypes of nrl−/− mutants. The nrl mutant zebrafish larvae have significantly increased levels of mRNA abundance compared to wildtype, consistent with the notion that nrl has auto-regulatory properties. Similarly, tbx2b mutants also have high nrl transcription, suggesting a repressive interaction. The nrl−/− and nrl−/−;tbx2b−/− mutants share consistently similar transcription profiles across most other assayed genes. Examination of the opsin genes expressed in rods (rho1) and UV cones (sws1) reveals disruption to expected levels of opsin relative to observed abundance of each cell type. Despite their observed overabundance of rods, tbx2b mutants have slightly lower levels of rho1 transcription. Similarly, despite nrl and nrl−/−/tbx2b−/− mutants having increased UV cone abundance, sws1 mRNA abundance does not show similar increases. Error bars represent standard error of the mean. N = 3 to 6 biological replicates. * = P < 0.05, ** = P < 0.005, *** = P < 0.0005 as determined by Brown-Forscythe and Welch 1-way ANOVA with Dunnett T3’s multiple comparison tests. |