- Title
-
CRK and NCK adaptors may functionally overlap in zebrafish neurodevelopment, as indicated by common binding partners and overlapping expression patterns
- Authors
- Stergas, H.R., Dillon-Martin, M., Dumas, C.M., Hansen, N.A., Carasi-Schwartz, F.J., D'Amico, A.R., Finnegan, K.M., Juch, U., Kane, K.R., Kaplan, I.E., Masengarb, M.L., Melero, M.E., Meyer, L.E., Sacher, C.R., Scriven, E.A., Ebert, A.M., Ballif, B.A.
- Source
- Full text @ FEBS Lett.
|
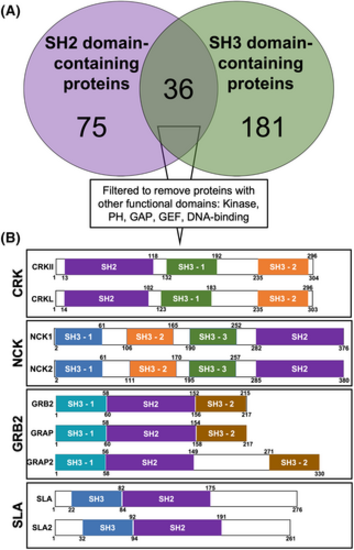
Bioinformatic analysis identifies four protein families comprised of only SH2 and SH3 domains. (A) Venn diagram of human SH2 domain-containing proteins (purple) and SH3 domain-containing proteins (green). (B) Removing proteins containing other functional domains leaves four adaptor protein families: CRK, NCK, GRB2, and SLA. Colors represent domains with greatest amino acid homology. Numbers correspond to amino acid residues |
|
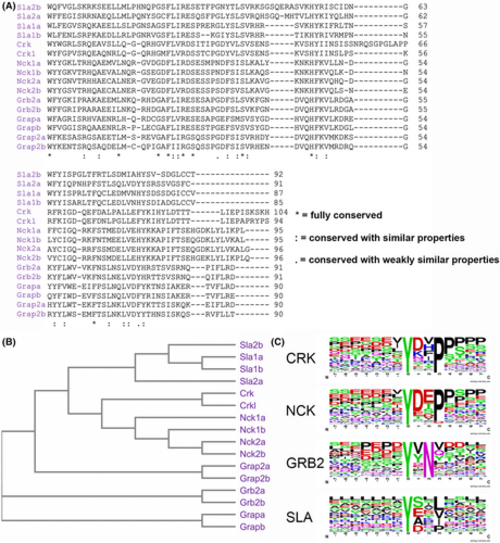
SH2 domain homology and binding motif comparison of Crk, Nck, Grb2, and Sla family adaptors. (A) Amino acid alignment of zebrafish SH2 domains from the Crk, Nck, Grb2, and Sla families. (B) Phylogenetic tree of the SH2 domains from the alignment in A. (C) Weblogos, centered around phosphotyrosine, depicting the preferred SH2 binding motifs of the indicated adaptor families. |
|
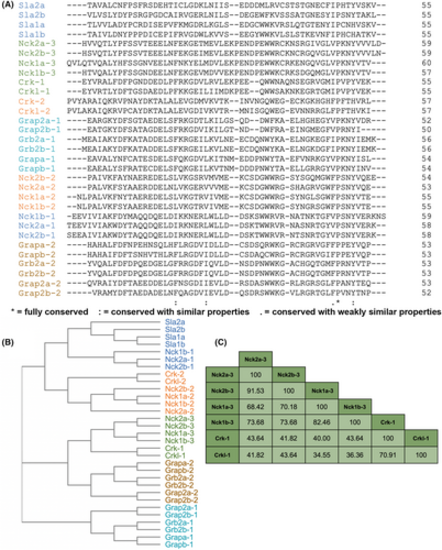
Homology comparison of SH3 domains from Crk, Nck, Grb2, and Sla family adaptors. (A) Amino acid alignment of zebrafish SH3 domains from the Crk, Nck, Grb2, and Sla families. Colors represent domains with greatest amino acid homology. (B) Phylogenetic tree of SH3 domains from alignment in A. (C) Percent identity table of Crk-SH3-1 and Nck-SH3-3 domains from the alignment in A. |
|
SH2 and SH3 domain-containing adaptors show overlapping expression in the zebrafish nervous system at 48 hpf. Wholemount lateral (A–J) and dorsal images (A′–J′) of in situ hybridization stained zebrafish embryos showing crk (A, A′), crkl (B, B′), grb2a (C, C′), grb2b (D, D′), nck1a (E, E′), nck1b (F, F′), nck2a (G, G′), nck2b (H, H′), sla1a (I, I′), and sla2b (J, J′) mRNA expression. Inset image (A–J) is a lateral view of a control-stained embryo using a corresponding sense probe. Scale bar in D and D′ is 250 μm. (K) RNAseq expression [18] of SH2 and SH3 domain-containing adaptors with high and low expression control genes (mob4 and tbp) from whole 48 hpf zebrafish. Expression is in transcripts per million (TPM). The gray box with diagonal lines indicates no detected expression. E, eye; FB, fin bud; MHB, midbrain–hindbrain boundary; OT, optic tectum; OV, otic vesicle. |
|
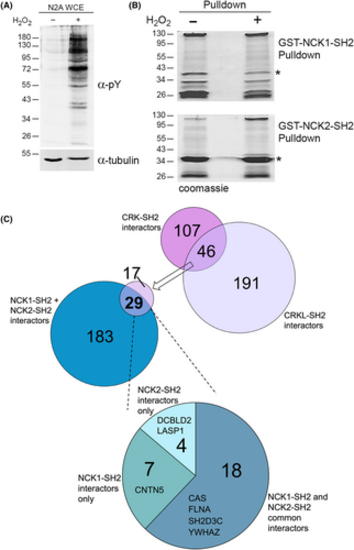
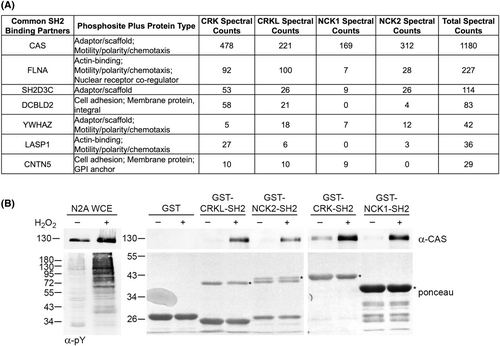
CRK and NCK family SH2 domains have overlapping neuronal binding partners. (A) Anti-phosphotyrosine (α-pY) and anti-tubulin western blots of extracts from Neuro2A cells treated with (+) or without (−) hydrogen peroxide (H2O2). Molecular weight markers in kilodaltons are indicated. (B) Representative coomassie-stained gels used for MS analysis of GST-NCK1-SH2 and GST-NCK2-SH2 pulldowns using extracts from A. *Indicates full-length GST-SH2 fusion (breakdown products are major bands below). (C) Previously [31], we found 46 proteins from Neuro2A cells were induced by H2O2 to bind to both the SH2 domains of CRK and CRKL. 29 of these were also found induced by H2O2 to interact with the SH2 domain of either NCK1 or NCK2. The pie chart indicates the proportions of the 29 proteins that interact with one or both NCK family member's SH2 domain. Binding partners involved in cell migration (see Fig. 6) are listed. |
|
CRK and NCK family SH2 domains have overlapping neuronal binding partners involved in cell migration. (A) The 29 common SH2-binding partners from Fig. 5 were categorized by molecular function and protein-type. Proteins that were categorized as cytoskeletal components, cell adhesion, adaptor/scaffold molecules, or motility/polarity/chemotaxis proteins were placed in the table. Also provided are total spectral counts from CRK-SH2 and NCK-SH2 MS. (B) CRK and NCK family SH2 domains are induced by H2O2 to interact with CAS, a major component of focal adhesions. Anti-CAS western blot of whole-cell extract (WCE) and the indicated pulldowns using extracts from Neuro2A cells treated with (+) or without (−) H2O2. The anti-pY blot shows effective stimulation by H2O2. *Indicates full-length GST-SH2 fusion (breakdown products are major bands below). Molecular weight markers in kilodaltons are indicated. |
|
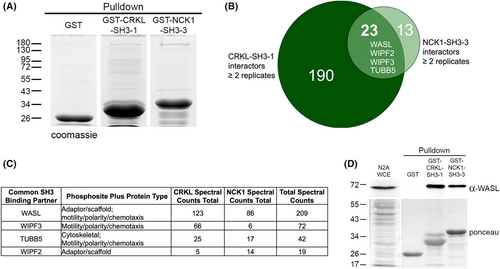
CRKL-SH3-1 and NCK1-SH3-3 share overlapping neuronal interacting partners involved in cell migration. (A) Representative coomassie-stained gels used for MS analysis of GST, GST-CRKL-SH3-1, and GST-NCK1-SH3-3 pulldown experiments using extracts from Neuro2A cells. Molecular weight markers in kilodaltons are indicated. (B) Comparison of CRKL-SH3-1 (dark green) and NCK1-SH3-3 (light green) interactors found in at least two replicates reveals 23 overlapping interactors. Binding partners involved in cell migration are shown in the overlap. (C) The 23 common SH3-binding partners were characterized by molecular function and protein-type. Proteins that were categorized as cytoskeletal components, adaptor/scaffold molecules, or motility/polarity/chemotaxis proteins were placed in the table. Also provided are total spectral counts from CRKL and NCK1 MS runs. (D) CRKL-SH3-1 and NCK1-SH3-3 both interact with WASL, an actin-binding protein involved in cell migration. Anti-WASL western blot of whole-cell extract (WCE) and GST, GST-CRKL-SH3-1, and GST-NCK1-SH3-3 pulldowns using Neuro2A lysate. Ponceau-stained membranes indicate lysate before pulldowns (left panel) and pulldowns (right panel). Molecular weight markers in kilodaltons are indicated. |
|
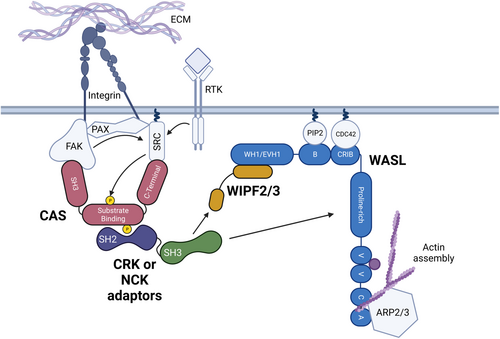
Model of CRK and NCK family overlapping function in cell motility and adhesion pathways. Activation of integrin receptors by binding to the extracellular matrix (ECM) or autophosphorylation of receptor tyrosine kinases (RTK) through ligand binding leads to the activation of SRC (arrows). Upon integrin receptor activation, Focal Adhesion Kinase (FAK), Paxillin (PAX), SRC, CRK-Associated Substrate of SRC (CAS), and other proteins cluster to form focal adhesions – Centers of adhesion and migratory signaling near the cell membrane. SRC primarily phosphorylates (yellow) CAS on tyrosine – X-X-proline motifs; CAS contains 15 Y-X-X-P motifs in its substrate binding domain. Tyrosine phosphorylation allows CRK or NCK family adaptor proteins to bind via their SH2 domain (purple). The CRK/NCK SH3 domain (green) binds either directly to WASL via its proline-rich domain or indirectly to WIPF2 or WIPF3. WASL's proximity to the focal adhesion drives localized Actin polymerization through the ARP2/3 complex. |