- Title
-
Molecular analyses of zebrafish V0v spinal interneurons and identification of transcriptional regulators downstream of Evx1 and Evx2 in these cells
- Authors
- England, S.J., Rusnock, A.K., Mujcic, A., Kowalchuk, A., de Jager, S., Hilinski, W.C., Juárez-Morales, J.L., Smith, M.E., Grieb, G., Banerjee, S., Lewis, K.E.
- Source
- Full text @ Neural Dev.
|
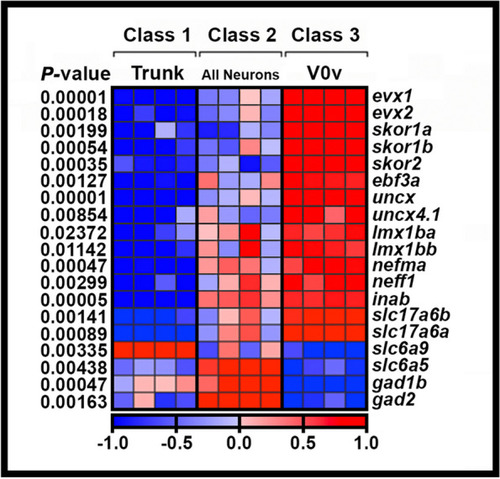
Transcriptional profiling of V0v spinal interneurons. Heatmap analysis of gene-expression profiling of 27 h V0v spinal cord interneurons. A three-class ANOVA analysis of differential expression was performed on different FAC-sorted populations of cells. Class 1: All trunk cells. Class 2: All post-mitotic spinal neurons. Class 3: V0v interneurons. Each column is a different biological replicate. Rows show relative expression levels for a single gene as normalized data transformed to a mean of 0, with standard deviation of + 1 (highly expressed, red) or -1 (weakly/not expressed, blue) sigma units. Adjusted |
|
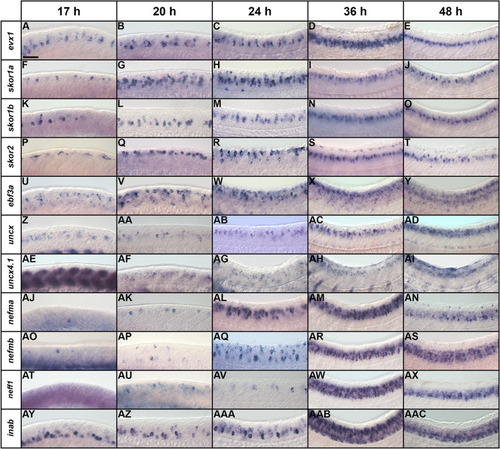
Temporal expression profiles of V0v candidate genes in zebrafish spinal cord. ( |
|
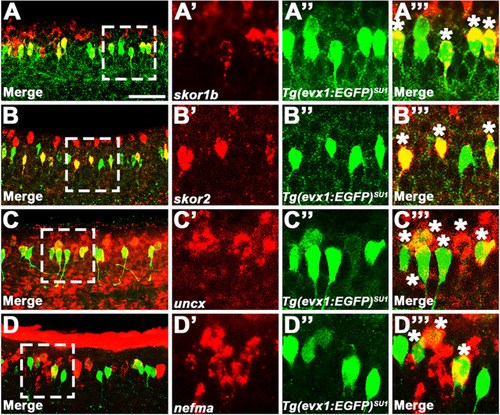
V0v candidate genes are co-expressed in subsets of V0v spinal interneurons. ( |
|
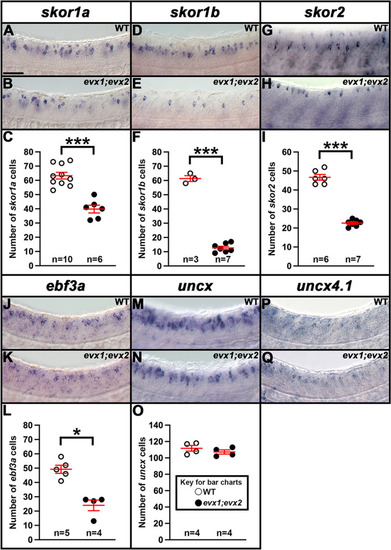
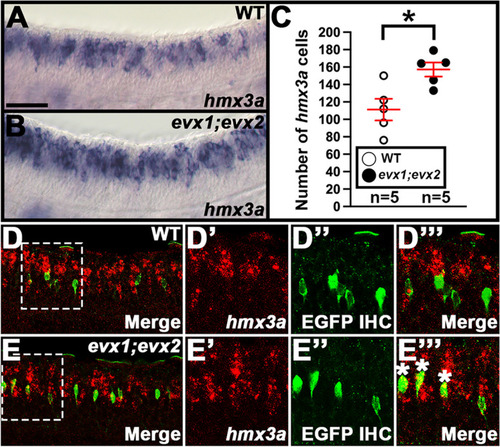
Expression of |
|
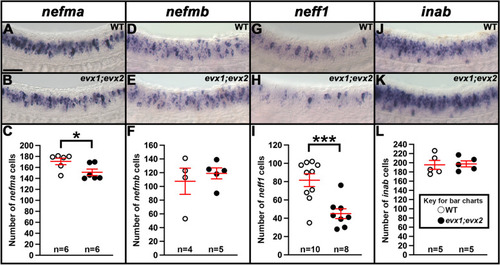
Expression of |
|
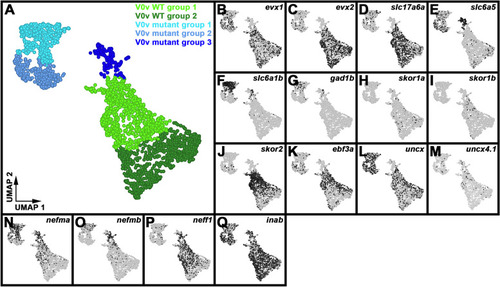
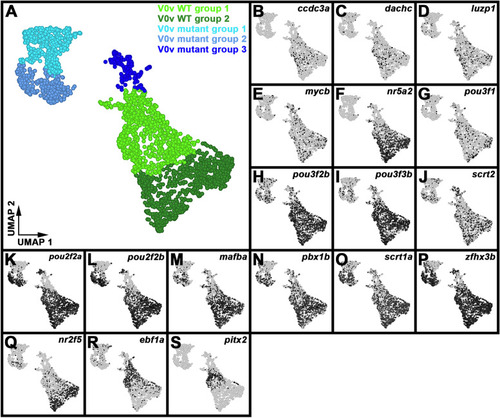
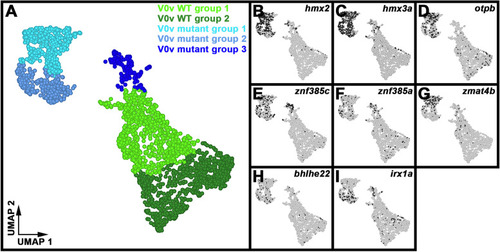
Single-cell RNA-seq analysis of WT and |
|
Differential gene expression identifies two distinct subsets of WT V0v spinal interneurons. ( |
|
|
|
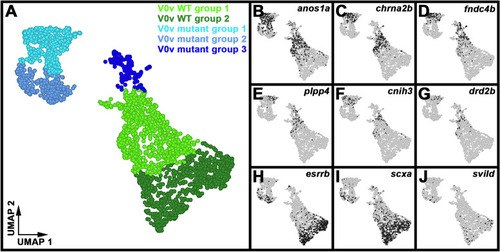
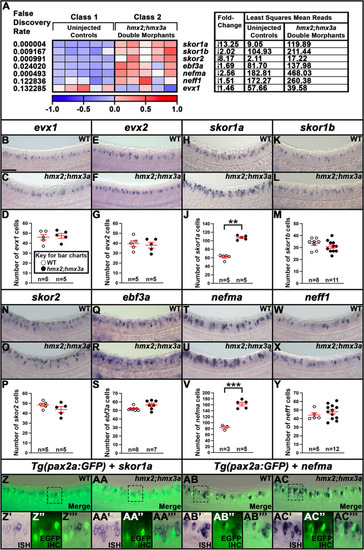
Genes downregulated in |
|
Genes upregulated in |
|
|
|
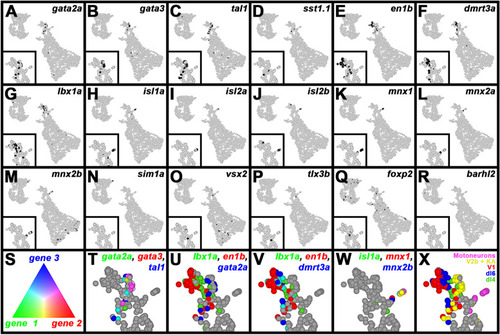
A subset of V0v spinal interneuron genes are upregulated in |
|
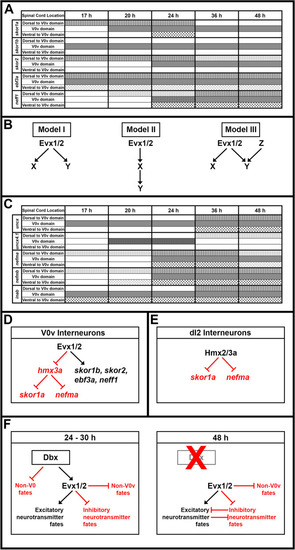
Possible GRNs downstream of Evx1/2 in V0v spinal interneurons. ( |