- Title
-
Role of microRNAs and their downstream target transcription factors in zebrafish thrombopoiesis
- Authors
- Al Qaryoute, A., Fallatah, W., Dhinoja, S., Raman, R., Jagadeeswaran, P.
- Source
- Full text @ Sci. Rep.
|
Knockdown screen of microRNAs and thrombocyte production. ( |
|
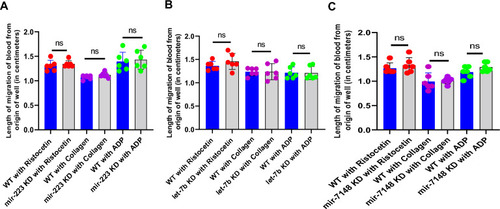
Effect of ( |
|
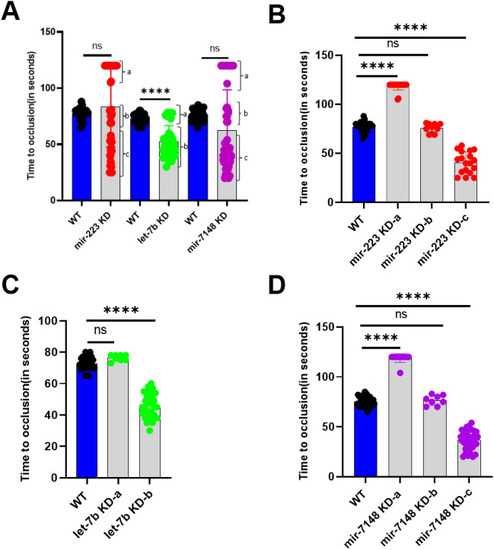
Arterial thrombosis in |
|
Levels of the downstream transcription factor mRNAs after knockdowns of ( |
|
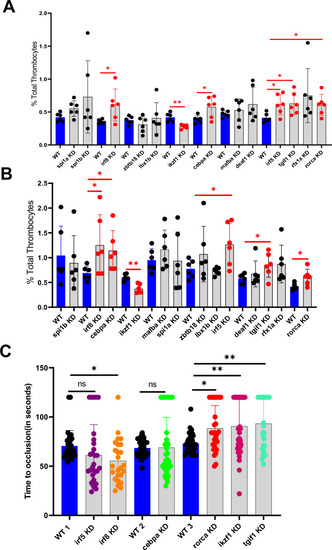
Flow cytometric analysis of total thrombocytes percentage in zebrafish whole blood. ( |
|
Functional evaluation of |