- Title
-
Knockdown of Yap attenuates TAA-induced hepatic fibrosis by interaction with hedgehog signals
- Authors
- Zhao, Y., Wang, H., He, T., Ma, B., Chen, G., Tzeng, C.
- Source
- Full text @ J. Cell Commun. Signal.
|
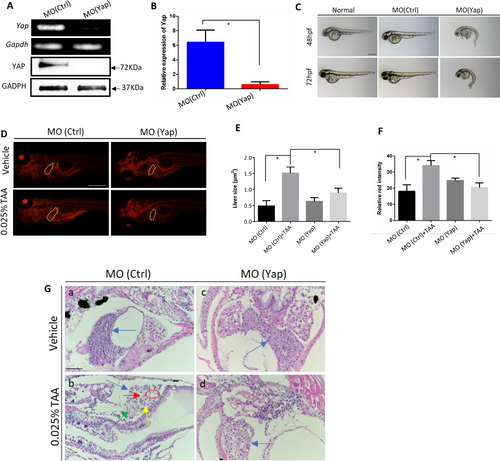
Analysis of morphant phenotype and TAA-induced embryonic liver lesions caused by Yap knockdown. A Validation of Yap knockdown by RT-PCR and Western blot analysis. Total RNA and proteins were extracted from 24 hpf zebrafish embryos in MO (Yap) and MO (Ctrl) (n = 10). B Validation of Yap knockdown by qPCR. C Morphant observation of 48 and 72 hpf embryos injected with MO (Yap) (5 ng/embryo), MO (Ctrl) (5 ng/embryo), and non-injected normal embryos. D Nile red-stained sections of embryos with the treatment of 0.025% TAA from 72 hpf to 7 dpf. E Qualifications of liver size in each group, n = 5. F Qualifications of Nile red staining by relative red intensity by ImageJ (n = 5). Data are shown as means ± SEM. *p < 0.05. G Histological evaluations of zebrafish liver by hematoxylin and eosin (H&E) with the treatment of 0.025% TAA from 72 hpf to 7 dpf |
|
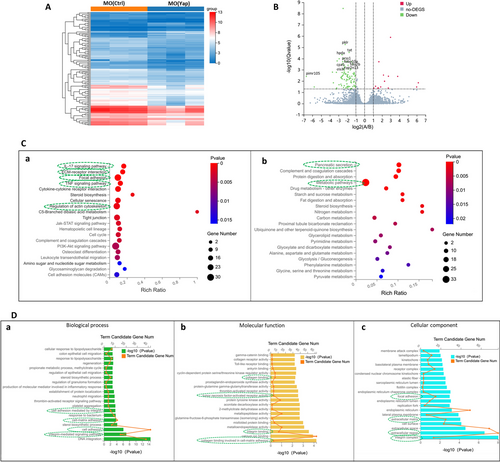
Global transcriptomic analysis and knockdown of Yap mediated signatures in zebrafish embryos. A Hierarchical clustering of RNA-Seq data shows global gene expression changes between the MO (Ctrl) and MO (Yap). High and low expression levels of genes are represented by red and blue. Gene expression (TPM) is log2-transformed and median-centered across genes. B Groups at mRNA level are shown in volcano plots of log2 (fold change) versus − log10 (p value). Red dots indicate up-regulated DEGs, the green dots indicate down-regulated DEGs, and blue dots represent non-DEGs. The top ten down-regulated transcripts, ranked by their distance to the base point are marked. C KEGG enrichment analysis of DEGs. Top 20 KEGG pathways among the down-regulated (a) or (b) up-regulated DEGs were presented in bar charts. D GO analysis of downregulated DEGs mediated by Yap knockdown. Top 20 GO in the three categories: biological process (a), molecular function (b) and cellular component (c) of significantly down-regulated DEGs were presented in bar charts |
|
Gene set enrichment analysis (GSEA) was performed using the Molecular Signatures Database. A Downregulated genes compared from MO (Yap) to MO (Ctrl) as signatures were enrichment by GSEA. B |
|
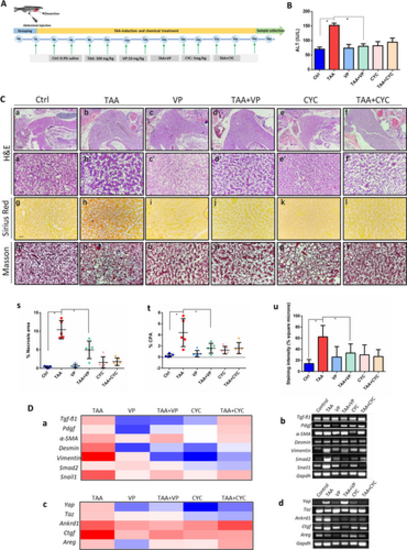
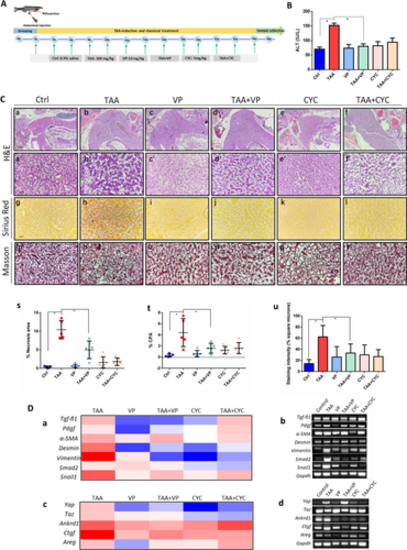
Assessment of liver histopathology. A The experimental flow chart for chemical treatment and time points of sampling. B Liver function tests by ALT assay. Statistical test used was one-tailed student t-test, *p < 0.05. C |
|
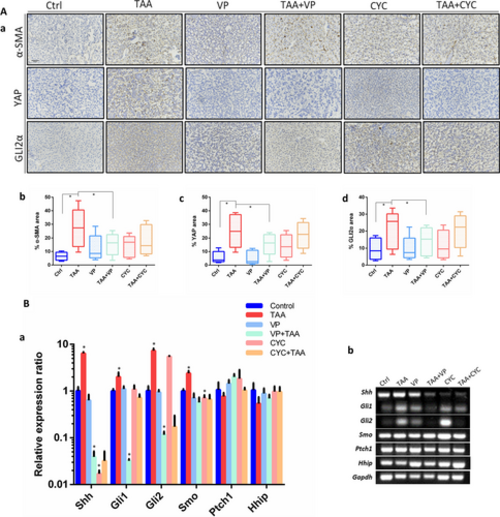
Immunohistochemistry detections in zebrafish liver. A (a) Immunostaining of α-SMA, YAP and GLI2α, for the detection of tissue sections of normal control, TAA-induced, VP treatment, TAA + VP treatment, CYC treatment and TAA + CYC treatment. (b–d |
|
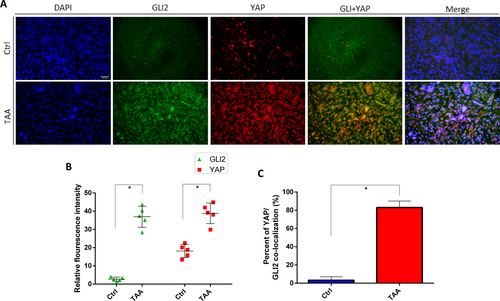
Immunohistochemical detection of the distribution of YAP and GLI2α in TAA-induced liver fibrosis. A Representative fluorescence microscopy of the distributions of YAP and GLI2α in liver sections using the indicated antibodies: anti-YAP (red), anti-GLI2α (green) and nuclei were stained with 4′, 6-diamidino-2-phenylindole DAPI (blue). The corresponding overlay of YAP and GLI2α is shown in panel (YAP + GLI2α) and the corresponding overlays of the three signals is shown in panel (merge). Scale bar = 50 µm. B Quantification of relative fluorescence intensity of YAP and GLI2α in control and TAA-induced liver fibrosis. C Percent of YAP/GLI2α nuclear colocalization in control and TAA-induced liver fibrosis. The fluorescence intensity of each section was measured by ImageJ. *p < 0.05, n = 5, means ± SD |
|
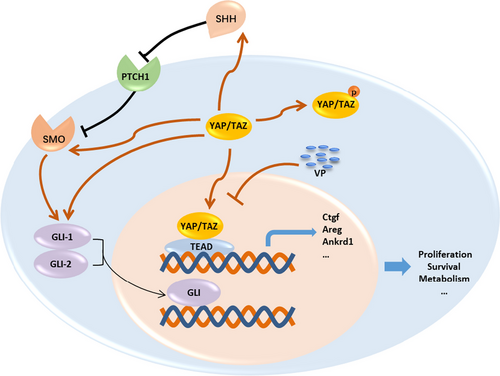
Crosstalk between Yap with Hh signaling factors. Upon Hippo pathway inactivation, activated YAP/TAZ translocate into the nucleus and interact with the TEAD family transcription factors, thereby by promoting the expression of target genes of YAP, including Ankrd1, Ctgf, Areg, etc. VP inhibits YAP through direct binding with its cofactor TAZ, thereby inhibiting the interactions with its downstream cofactors, TEAD. Inhibition of Yap exhibited suppression effect on TAA-induced Shh, Smo, Gli1 and Gli2. YAP yes-associated protein, TAZ transcriptional coactivator with PDZ-binding motif, VP verteporfin, Smo smoothened, SHH sonic hedgehog, PTCH1 patched 1, Gli GLI family zinc finger, Ctgf connective tissue growth factor, Areg amphiregulin; connective tissue growth factor, Ankrd1 ankyrin repeat domain 1 |