- Title
-
Comprehensive Transcriptome Analysis Reveals Sex-Specific Alternative Splicing Events in Zebrafish Gonads
- Authors
- Lin, X., Liu, F., Meng, K., Liu, H., Zhao, Y., Chen, Y., Hu, W., Luo, D.
- Source
- Full text @ Life (Basel)
|
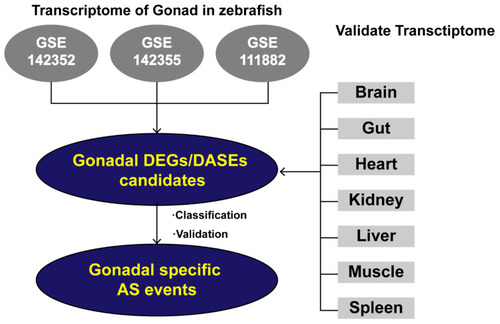
Analytical workflow. Three independent sets of RNA-seq data (GSE142352, GSE142355, GSE111882) from matched zebrafish testes and ovaries were used to conduct genome-wide analysis of gonadal expression levels and AS events in female and male zebrafish. The DEGs and DASGs shared by the three data sets are referred to as gDEGs and gDASGs. Following this, the gDASGs were compared with RNA-Seq data (PRJNA690124) from brain, gut, heart, kidney, liver, muscle, spleen to obtain gonadal specific gDASGs. |
|
Number of DEGs and DASGs in zebrafish and chromosome distribution profile for clusters of gDEGs and gDASGs (A) Bar graph representing the statistics of DEGs highly expressed in ovary and testis respectively. (B) Relative chromosome distributions in percentages of gDEGs in testis and ovary. (C) Venn diagram of DASGs at different datasets. (D) Distribution of gDASGs on chromosomes. |
|
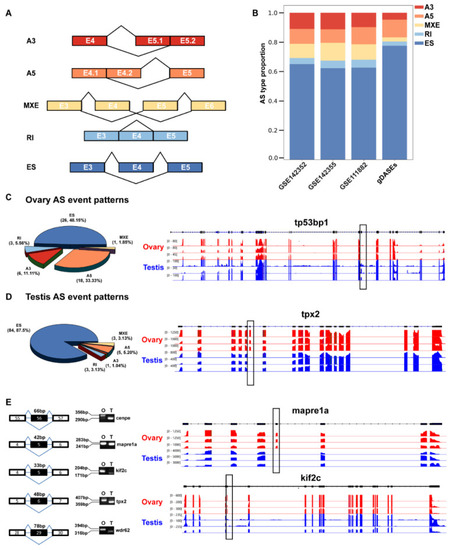
Distribution of AS types and in frame ratio in male and female zebrafish gonad. (A) Splice model for splice types detected in the rMATS projects. Exon number was illustrated in the box. (B) Proportion of the identified DASEs. ES, exon skipping; A5SS, alternative 5′ splice site; A3SS, alternative 3′ splice site; MXE, mutually exclusive exon; RI, retained intron; (C) Proportion of the gDASEs in ovary and case. (D) Proportion of the gDASEs in testis and case. (E) The differentially spliced transcripts of mapre1a, tpx2, cenpe, kif2c and wdr62 were validated by RT-PCR using cDNA samples from adult (6 mpf) zebrafish gonads (O for ovaries and T for testes). The RNA-seq tracks of tp53bp1, tpx2, mapre1a, and kif2c are shown in the right panel. The differentially spliced exons between ovaries and testes are indicated with boxes. |
|
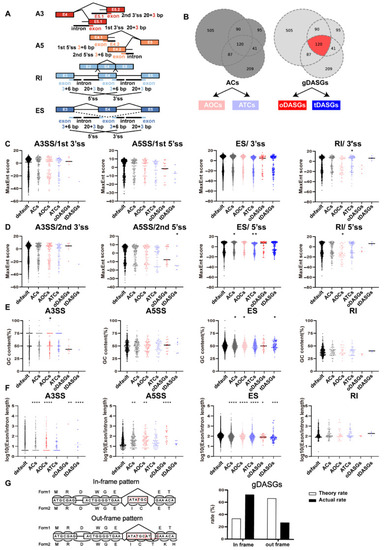
Sequence characteristics of the AS exons and introns in ovaries and testes. (A) Models of splicing related SNPs in different regions for A3SS, A5SS, ES and RI separately. SNPs were classified by position as four mutually exclusive regions, including splice (3′ss and 5′ss), exon (alternative fragment), gene (the splicing gene) and chromosome (out of the splicing gene). (B) model of different groups of DASGs (ACs:All Candidates, AOCs: All Ovary Candidates, ATCs: All Testis Candidates, oDASGs: Ovary specific gDASGs, tDASGs: Testis specific gDASGs). (C,D) MaxEnt score of 5′ss and 3′ss, (E) GC content and (F) length of alternative fragment in splice type A3SS, A5SS, ES and RI, separately.(G) model of frame-preserving or frame-switching and theory rate and actual rate of gDASGs. Exon length determines whether an alternatively spliced single-exon skip is frame-preserving or frame-switching. We define an alternatively spliced exon as frame-preserving if its length is an exact multiple of 3 nt, as its alternative splicing will not alter the protein reading frame of subsequence exons (top). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. |
|
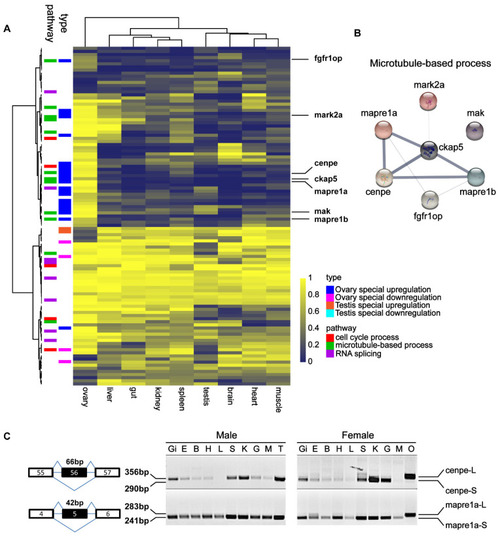
Identification of testis- and ovary-specific AS genes using multi-tissue RNA-Seq data. (A) Heatmap of PSI value of ovary, liver, gut, kidney, spleen, testis, brain, heart and muscle. (B) Protein-protein interaction network. (C) Differentially expressed splicing transcripts of cenpe and mapre1a were validated by RT-PCR in gonads and other tissues from adult (6 mpf) male and female zebrafish. The DNA fragments were sequenced and shown in Figure S6. ‘Gi’: gill; ‘E’: eye; ‘B’: brain; ‘H’: heart; ‘L’: liver; ‘S’: spleen; ‘K’: kidney; ‘G’: gut; ‘M’: muscle; ‘T’: testis; and ‘O’: ovary. |
|
Cases for the biological significances of the sex specific AS events. (A) The differentially spliced transcripts of ncoa5 were validated in adult (6 mpf) zebrafish gonads (O for ovaries and T for testes) by RT-PCR. (B) Multiple protein sequence alignment of RRM1 from Tachysurus fulvidraco (T.f.; XP_027002460.1), Danio rerio (D.r.; XP_005166217.1), Cyprinus carpio (C.c.; XP_018925131.1), Carassius auratus (C.a.; XP_026121940.1), Salmo salar (S.s.; NP_001133374.1), Cynoglossus semilaevis (C.s.; XP_024915677.1), Oryzias latipes (O.l.; XP_023810720.1), Paralichthys olivaceus (P.o.; XP_019952347.1), Oreochromis niloticus (O.n.; XP_019214575.1) and Monopterus albus (M.a.;XP_020442329.1) obtained with Clustal Omega, the skip region is marked by red box (grey bottom). The asterisk (*) indicates the identical amino acid residue. (C) 3D protein structure modeling of ncoa5-S and ncoa5-L, with the RRM domain represented by a blue ellipse and the skip region designated by a red box, both of which contain sheet and helical structures. |