- Title
-
Phenolic Lipids Derived from Cashew Nut Shell Liquid to Treat Metabolic Diseases
- Authors
- Sahin, C., Magomedova, L., Ferreira, T.A.M., Liu, J., Tiefenbach, J., Alves, P.S., Queiroz, F.J.G., Oliveira, A.S., Bhattacharyya, M., Grouleff, J., Nogueira, P.C.N., Silveira, E.R., Moreira, D.C., Leite, J.R.S.A., Brand, G.D., Uehling, D., Poda, G., Krause, H., Cummins, C.L., Romeiro, L.A.S.
- Source
- Full text @ J. Med. Chem.
|
Figure 1. Chemical structures of fibrates, thiazolidinediones (TZDs), and glitazars. |
|
Figure 2. Similarity of chemical structures of stearic acid and saturated anacardic acid. |
|
Figure 3. Saturated derivatives (3–23) designed from the mixtures of anacardic acids (1) and cardanols (2). |
|
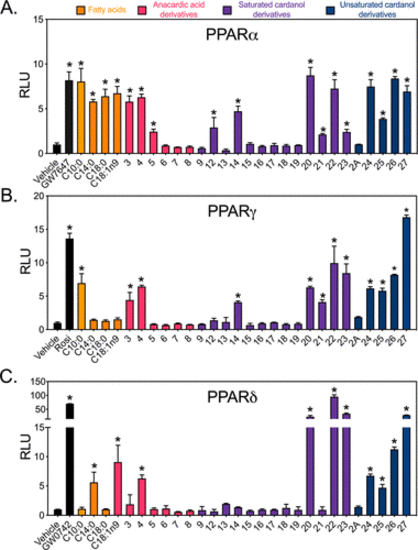
Figure 4. In vitro screening of CNSL derivatives for PPARα, PPARγ, and PPARδ activity reveals a subset of selective pan-activators. HEK293 cells were transiently co-transfected with GAL4-hPPARα (A), GAL4-hPPARγ (B), or GAL4-hPPARδ (C) together with UAS-luciferase reporter and treated with positive controls (10 nM GW7647, 100 nM Rosi, and 10 nM GW0742) or 50 μM of indicated compounds for 16 h. Data represent mean ± standard deviation (SD) (N = 3). RLU, relative luciferase units = luciferase light units/β-galactosidase × time. Vehicle (DMSO) response was set to 1. C10:0, decanoic acid; C14:0, myristic acid; C18:0, stearic acid; C18:1n9, oleic acid. *P < 0.05 relative to corresponding vehicle, using one-way analysis of variance (ANOVA) with Holm–Šidák correction. |
|
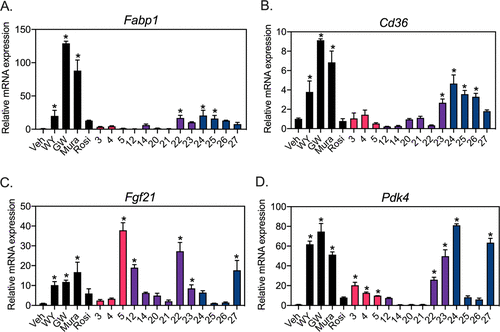
Figure 5. CNSL derivatives activate PPARα target genes in primary hepatocytes in a gene-selective manner. Primary hepatocytes were isolated from wild-type (WT) mice and incubated with vehicle (Veh, DMSO), 50 μM CNSL derivatives or 10 μM of positive controls: WY14643 (WY, PPARα agonist), GW7647 (GW, PPARα agonist), muraglitazar (Mura, PPARα/γ agonist), rosiglitazone (Rosi, PPARγ agonist) for 16 h. Expression of fatty acid uptake genes Fabp1 (A) and Cd36 (B), and fatty acid oxidation genes, Fgf21 (C) and Pdk4 (D) were analyzed by quantitative polymerase chain reaction (QPCR). Veh mRNA expression was set to 1. Data represent mean ± SD (N = 3). *P < 0.05 vs Veh using one-way ANOVA with Holm–Šidák correction. |
|
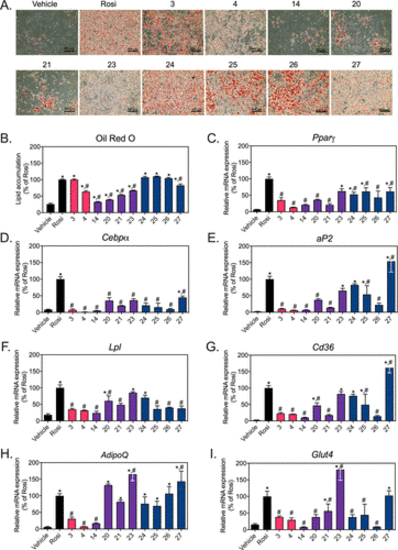
Figure 6. CNSL derivatives differentially regulate the expression of PPARγ target genes and adipocyte differentiation in 3T3-L1 cells. 3T3-L1 fibroblasts were differentiated for 11 days in the presence of vehicle (DMSO), 25 μM of indicated CNSL derivatives or 10 μM rosiglitazone (Rosi). Cells were harvested for Oil Red O staining and mRNA on day 11. (A) Cells were imaged under 10× magnification (n = 2/per group). (B) Lipid accumulation was quantitated by spectrophotometric analysis of extracted Oil Red O. mRNA expression was analyzed by QPCR for two key regulators of adipogenesis, (C) Pparγ and (D) Cebpα; fatty acid uptake genes (E) aP2 (Fabp4), (F) Lpl, and (G) Cd36; adipose-specific adipokine gene (H) AdipoQ and glucose uptake gene (I) Glut4. Vehicle mRNA expression was set to 1 and Rosi value was set to 100%. Data represent mean ± SD (N = 3). *P < 0.05 vs vehicle and #P < 0.05 vs Rosi; using one-way ANOVA with Holm–Šidák correction. |
|
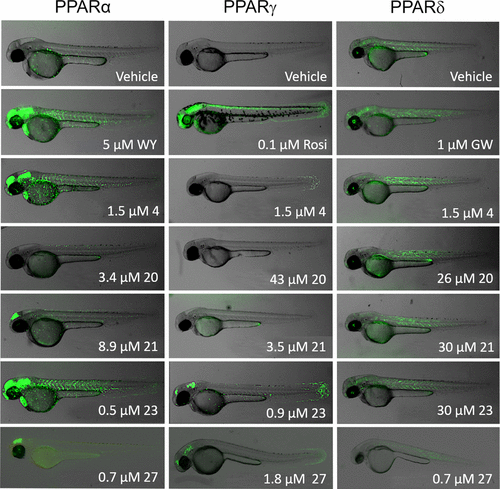
Figure 7. In vivo testing of CNSL derivatives using transgenic zebrafish that express human PPARα, PPARγ, or PPARδ reveal tissue-specific activation. Activation of human PPAR in the zebrafish embryo results in GFP expression. Basal activity of PPARα, PPARγ, and PPARδ is observed with vehicle (DMSO) treatment and is strongly increased in the presence of the full agonist for each receptor. Positive controls are WY (WY14643) for PPARα, Rosi (rosiglitazone) for PPARγ, and GW (GW0742) for PPARδ, respectively. Compounds were screened at their respective EC50’s determined from their dose–response curves in HEK293 cells with a few exceptions: 4 was screened at 1.5 μM for all receptors because of toxicity at higher concentrations; for PPARδ, 20 and 27 were screened below their EC50’s due to toxicity at higher concentrations. Each image depicts a representative embryo. Note that embryos incubated with 27 were imaged using a different microscope. |
|
Figure 8. Thermostabilization of hPPARα-LBD, hPPARγ-LBD, and hPPARδ-LBD by carboxylic acid-containing CNSL derivatives. Thermal shift assays of hPPARα-LBD (A) interacting with 25 μM GW7647 or 25 μM CNSL derivatives. Stabilization of the hPPARγ-LBD (B) and the hPPARδ-LBD (C) by 50 μM Rosi (rosiglitazone), 50 μM CAY (CAY10592), or 50 μM CNSL derivatives, respectively. The temperature–response curves were analyzed using the four-parameter dose–response curve function in GraphPad Prism 8 and Tm values were determined. ΔTm values were calculated for each compound relative to DMSO and are listed in the legend. Data points represent mean ± standard error of the mean (SEM) (N = 3). |
|
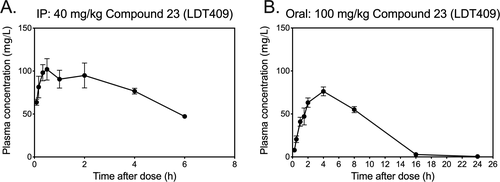
Figure 9. In vivo pharmacokinetic profile of 23 (LDT409) in C57BL/6 mice. The plasma concentration of 23 (LDT409) in mice after (A) a single intraperitoneal injection (IP) at 40 mg/kg and (B) oral administration in peanut butter treat at 100 mg/kg. Data represent mean ± SEM (N = 4 per time point). |