- Title
-
Genome Editing in Zebrafish by ScCas9 Recognizing NNG PAM
- Authors
- Liu, Y., Liang, F., Dong, Z., Li, S., Ye, J., Qin, W.
- Source
- Full text @ Cells
|
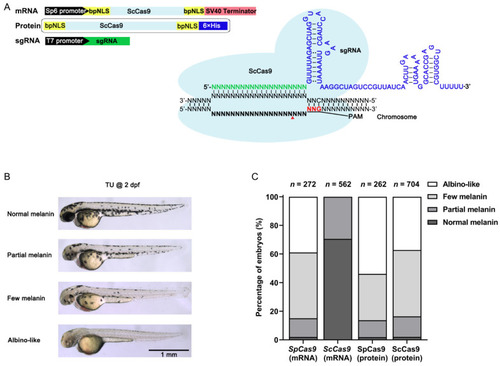
ScCas9 mRNA and protein induced tyr gene editing in zebrafish. (A), Schematic illustrating the ScCas9 system used in zebrafish. The ScCas9 system consists of two components, a dual bpNLS-tagged zebrafish codon-optimized ScCas9 protein and a sgRNA comprising a 20-nt seed sequence (green) infused with a sgRNA scaffold (blue). The ScCas9-bpNLS mRNA was transcribed in vitro from Sp6 promoter and sgRNA was transcribed from T7 promoter. ScCas9 protein tagged with bpNLS and His-tag was induced expression in vitro. Red triangle indicates the double stranded break sites induced by Cas9. bpNLS, bipartite nuclear localization signal; His, histidine tag. (B), Phenotypic evaluation of embryo pigment levels induced by targeting tyr using ScCas9 according to the amount of melanin. (C), Statistics of gene editing efficiency in the tyr target site induced by SpCas9 mRNA, ScCas9 mRNA, SpCas9 protein and ScCas9 protein. |
|
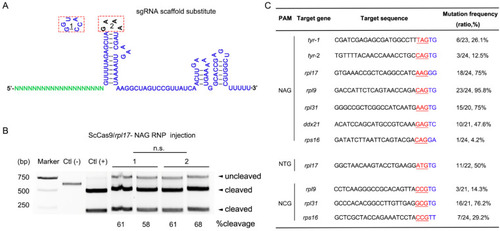
Genome editing in zebrafish using ScCas9. (A), Stem-loop differences between the ScCas9 original sgRNA tetraloop (1) and SpCas9 sgRNA tetraloop (2). (B), T7E1 assay of ScCas9/rpl17-NAG RNP-induced indel mutations. BsrBI-digested fragments of the purified PCR products were used as Ctl (+) and the undigested ones as Ctl (−). (C), Mutation frequency of ScCas9 at NNG PAM sites in zebrafish. The PAM is marked in red and underlined, and the downstream sequence of the PAM is marked in blue. |
|
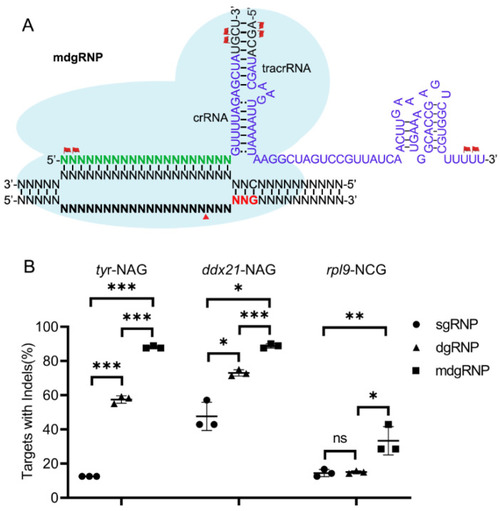
Chemically modified crRNA and tracrRNA duplex increased the ScCas9 efficiency. (A), Schematic illustration of the ScCas9 mdgRNP complex. The crRNA and tracrRNA scaffold with MS modified nucleotides (flag) at both termini coupled with the ScCas9 protein were loaded to the chromosome. (B), Statistics of gene editing efficiency induced by sgRNP, dgRNP and mdgRNP. ns, no significant difference; * p value < 0.05; ** p value < 0.01; *** p value < 0.001. |