- Title
-
A hybrid of light-field and light-sheet imaging to study myocardial function and intracardiac blood flow during zebrafish development
- Authors
- Wang, Z., Ding, Y., Satta, S., Roustaei, M., Fei, P., Hsiai, T.K.
- Source
- Full text @ PLoS Comput. Biol.
|
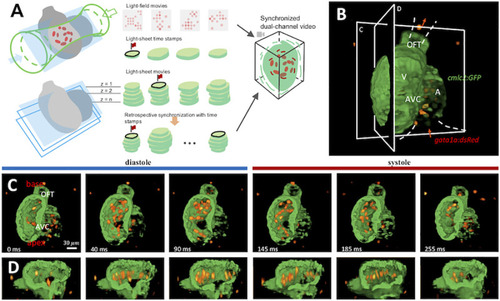
(A) The integration of light-sheet and light-field microscopy captures the contracting myocardium ( |
|
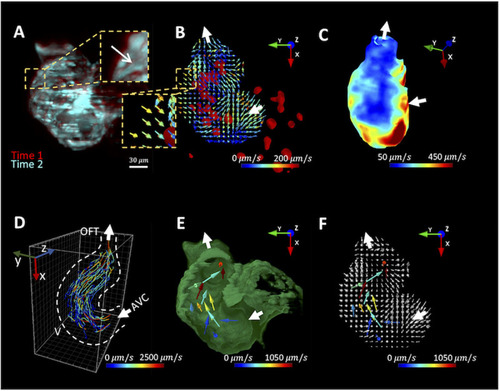
(A) Images of the myocardium from 2 different time points are overlaid to demonstrate the myocardial displacement. The arrow indicates the direction of displacement. (B) Deformable image registration (DIR) is used to infer the myocardial displacement between frames. Each vector indicates the direction and magnitude of the displacement velocity as color-coded by the magnitude ( |
|
(A) The trajectory of the blood cells is depicted as the flow centerline, starting from the AV canal (dark colors) to the outflow tract (light colors). (B) The myocardium is divided into 4 segments to assess regional variations in the vector field. (C) The velocity of individual blood cells is projected onto the flow centerline, and the mean flow velocity is plotted as a function of time. Myocardial velocity in each segment is quantified as the magnitude-weighted mean vector for each segment, as plotted in the color line. The black dotted line demonstrates the average velocity magnitude of the entire ventricle. (D) Six representative time points (from t1 to t6) of the heatmaps are presented for the myocardium (upper panels, color-coded in vector magnitude) and vector maps (lower panels). Red arrows indicate the rotational direction of the mean vector for each segment. |
|
(A) Blood flow velocity is depicted during a cardiac cycle. To compare among different heart rates, we normalize the time to one cardiac cycle for each zebrafish embryo. (B) The ventricle is divided into 4 segments. Arrows indicate the flow direction. The dotted line delineates the atrium. (C) The tracing of the myocardial velocity is normalized to one cardiac cycle in each segment. (D) The mean of relative displacement velocity is depicted in each segment during systole and diastole. The relative velocity in each segment is compared as the ratio of the segmental velocity to the mean of global velocity during a cardiac cycle. Kruskal-Wallis test and Dunn’s multiple comparisons test are performed in GraphPad Prism (** |