- Title
-
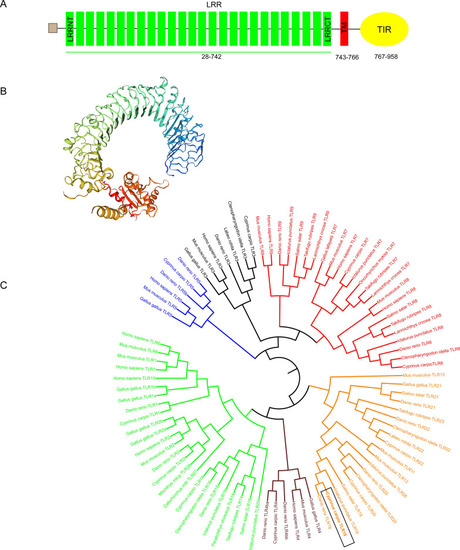
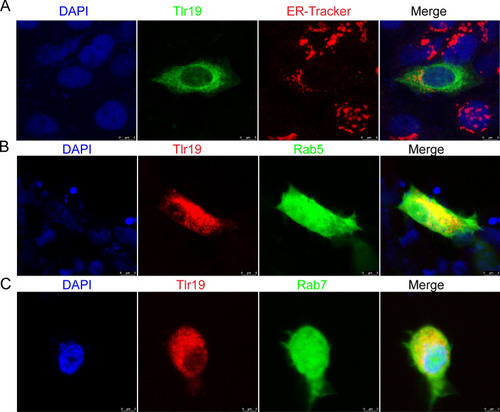
Identification and functional characterization of a fish-specific tlr19 in common carp (Cyprinus carpio L.) that recruits TRIF as an adaptor and induces ifn expression during the immune response
- Authors
- Shan, S., Liu, R., Feng, H., Meng, F., Aizaz, M., Yang, G.
- Source
- Full text @ Vet. Res.
|
|
|
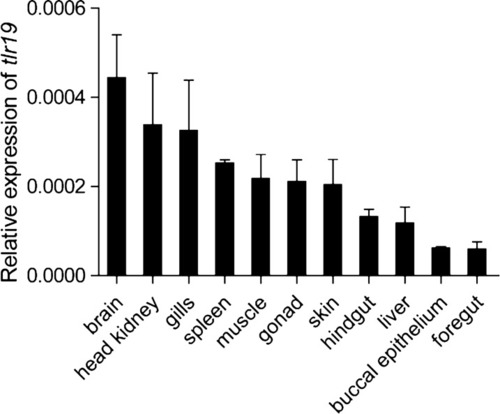
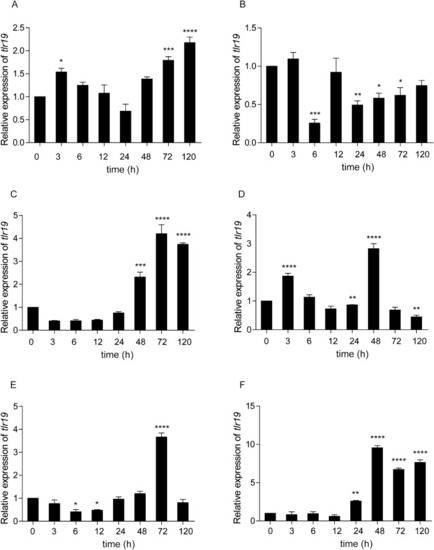
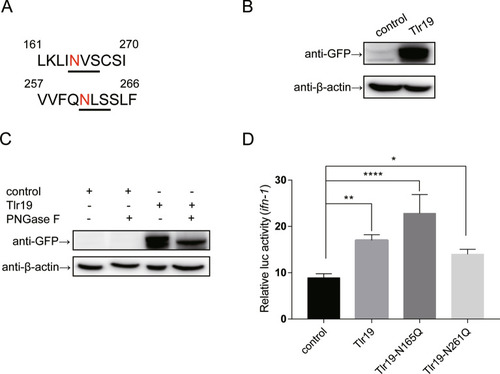
Tissue expression of Cctlr19 in normal common carp. The expression of Cctlr19 mRNA in the liver, spleen, head kidney, foregut, hindgut, skin, gills, gonad, muscle, buccal epithelium and brain was detected by qPCR. 40S ribosomal protein S11 in each tissue was amplified as an internal control, n = 3. |
|
|
|
|
|
|
|
|
|
|
|
|
|
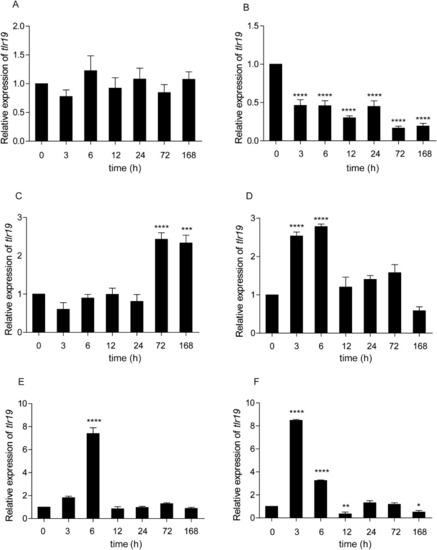
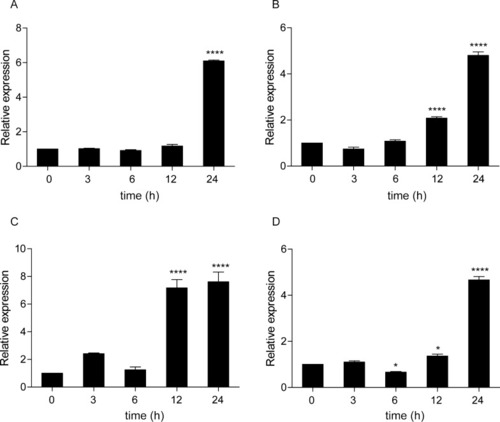
CcTlr19 induces the expression of cytokines. CcTlr19 was overexpressed in EPC cells in 24-well plates, and RNA was extracted using TRIzol. qPCR was used to test the expression of ifn-1 and viperin. The results were calculated relative to the expression of β-actin using qPCR. Mean ± SD (n = 3), **P < 0.01, ***P < 0.001, t test. |
|
|