- Title
-
Exposure to ethanol leads to midfacial hypoplasia in a zebrafish model of FASD via indirect interactions with the Shh pathway
- Authors
- Sidik, A., Dixon, G., Buckley, D.M., Kirby, H.G., Sun, S., Eberhart, J.K.
- Source
- Full text @ BMC Biol.
|
Ethanol interacts with PHENOTYPE:
|
|
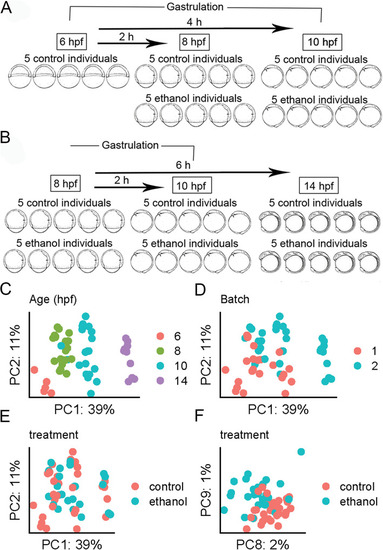
Developmental age is the dominant driver of variation in the dataset. a Schematic representation of the RNA-seq experimental design. Wild-type AB embryos were exposed to 1% ethanol at shield stage (6 hpf). Embryos were subsequently collected at 8 and 10 hpf for experiment 1 and b 8, 10, and 14 hpf, for experiment 2. c–f Principal components analysis (PCA) of the top 25,000 most variable genes. The percentage of variance explained is given on each axis label. c PC1 and PC2 color coded by developmental age. d PC1 and PC2 color coded by RNA-seq experiment (batch). e PC1 and PC2 color coded by ethanol treatment group. f PC8 and PC9 showing separation of ethanol-treated and control samples |
|
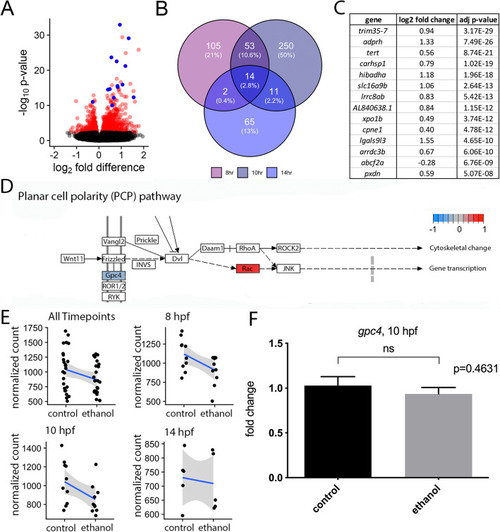
Effects on transcription are largely distinct between developmental time points. EXPRESSION / LABELING:
|
|
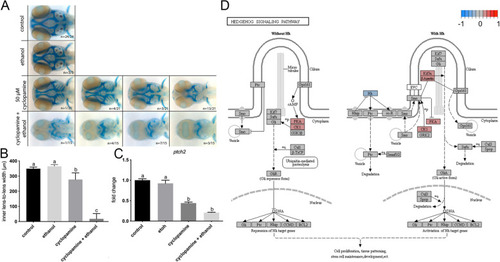
Ethanol indirectly attenuates Shh signaling. |
|
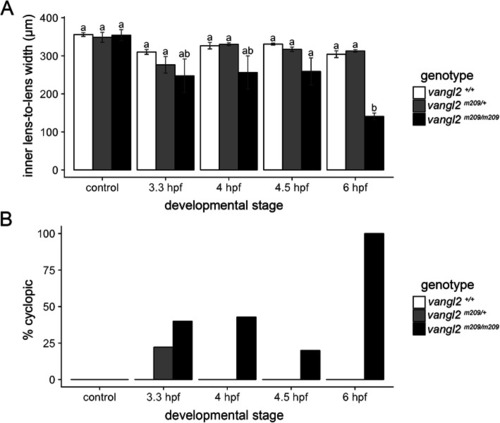
Ethanol disrupts convergent extension. PHENOTYPE:
|
|
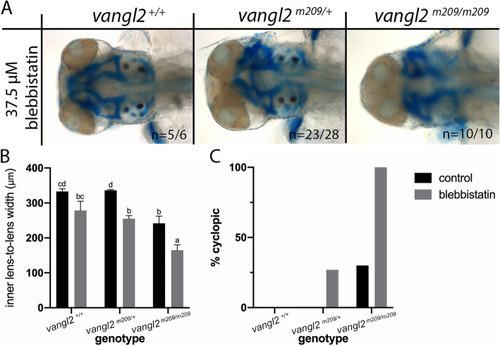
Blebbistatin phenocopies ethanol treatment in PHENOTYPE:
|
|
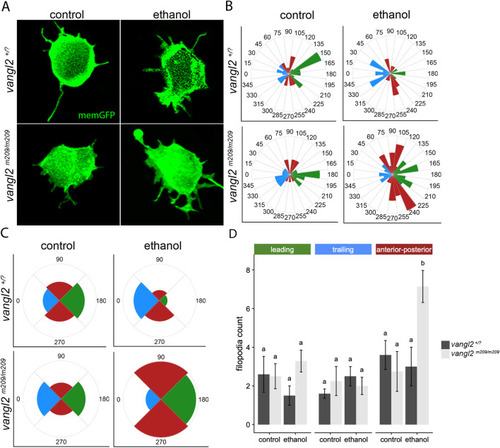
Ethanol disrupts the polarity of membrane protrusions in PHENOTYPE:
|