- Title
-
Three-dimensional collagen-based scaffold model to study the microenvironment and drug-resistance mechanisms of oropharyngeal squamous cell carcinomas
- Authors
- Miserocchi, G., Cocchi, C., De Vita, A., Liverani, C., Spadazzi, C., Calpona, S., Di Menna, G., Bassi, M., Meccariello, G., De Luca, G., Campobassi, A., Maddalena Tumedei, M., Bongiovanni, A., Fausti, V., Cotelli, F., Ibrahim, T., Mercatali, L.
- Source
- Full text @ Cancer Biol Med
|
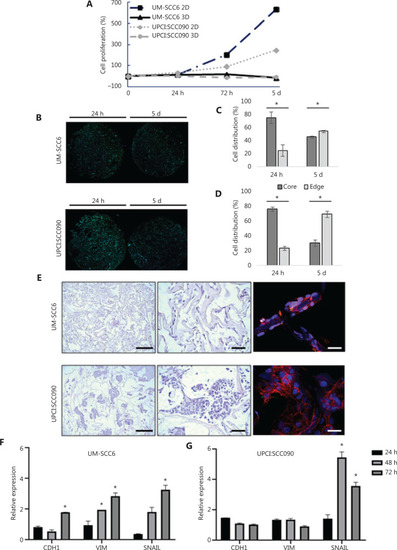
Characterization of the 3D oropharyngeal squamous cell carcinoma culture model. (A) Fold changes in percentage of cell proliferation (relative to hour 0) of UM-SCC6 and UPCI:SCC090 in monolayer (2D) and in scaffolds (3D) after 24 h, 72 h, and 5 days. (B) Whole images of histological sections of scaffold cultured with UM-SCC6 and UPCI:SCC090 on days 1 and 5. Cells are stained with DAPI (blue) and green is the collagen scaffold autofluorescence. Scale bars: 1 mm. (C, D) Cell percentage in the core and at the edge of the scaffold area. Data represent mean ± standard deviation. * |
|
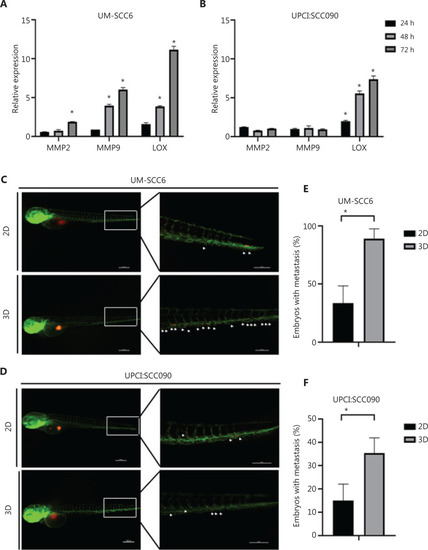
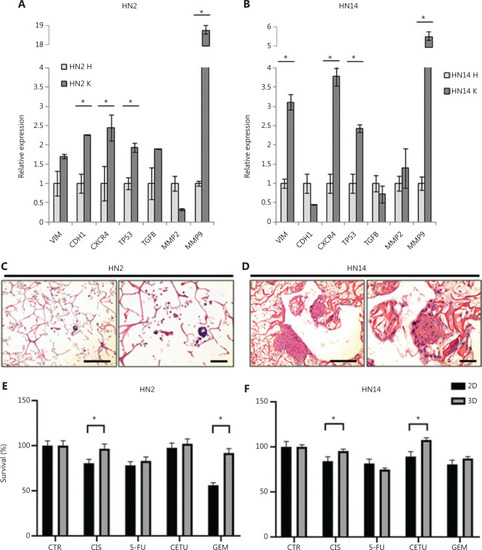
Cells cultured in the scaffold interact with collagen fiber network and display higher cell migration inside the vessels of zebrafish embryos. (A, B) UM-SCC6 and UPCI:SCC090 relative expression levels of |
|
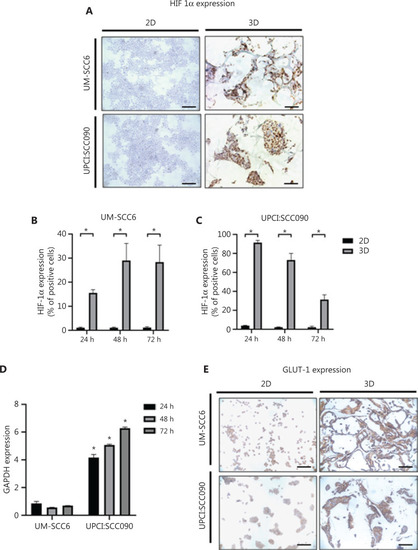
3D microenvironment induces hypoxic and glycolytic adaptation by oropharyngeal squamous cell carcinoma cells. (A) Representative images of HIF-1α-stained cytospinned cells and histological sections of 2D and 3D cultures (scale bar: 50 µm). (B, C) Percentage of HIF-1α-positive cells in 2D and 3D cultures of UM-SCC6 and UPCI:SCC090 after 24, 48, and 72 h. Data represent mean ± standard deviation. * |
|
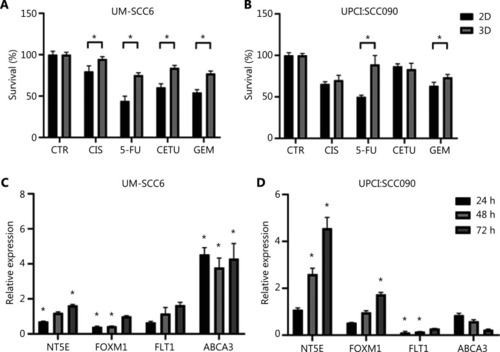
3D cultured cells acquire drug-resistance mechanisms. (A, B) Cytotoxicity analysis of UM-SCC6 and UPCI:SCC090 cell lines treated with cisplatin, 5-fluorouracil, cetuximab, and gemcitabine. Differences between 2D and 3D cultures were assessed by a 2-tailed Student’s |
|
Drug-resistance mechanisms are also conserved in HNC patient-derived primary cultures. (A, B) |