- Title
-
Quantification of cell behaviors and computational modelling show that cell directional behaviors drive zebrafish pectoral fin morphogenesis
- Authors
- Dokmegang, J., Nguyen, H., Kardash, E., Savy, T., Cavaliere, M., Peyriéras, N., Doursat, R.
- Source
- Full text @ Bioinformatics
|
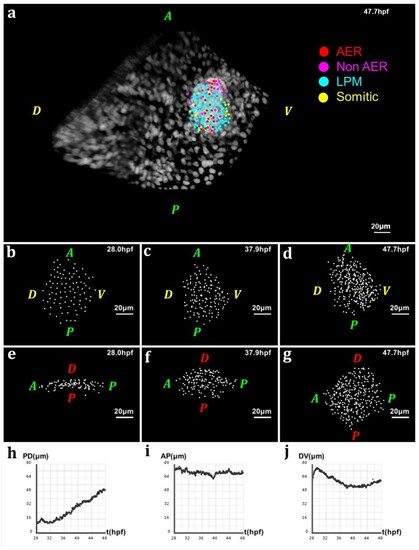
Geometry of the pectoral fin based on live imaging and image processing data. (a) 3D rendering of raw data nuclear staining at |
|
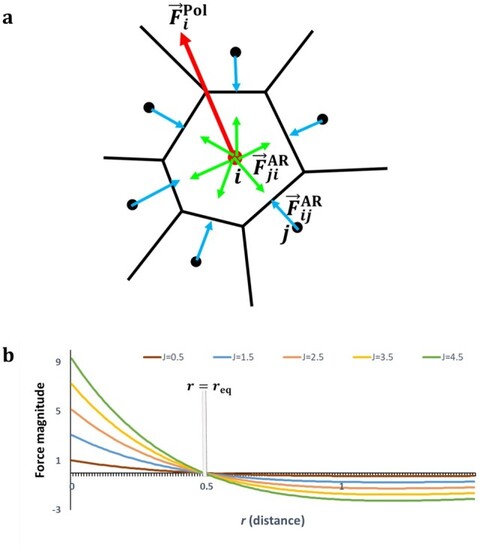
Center-based computational model of multicellular dynamics. (a) Schema of a local cell neighborhood and the abstract forces on cell centers. |
|
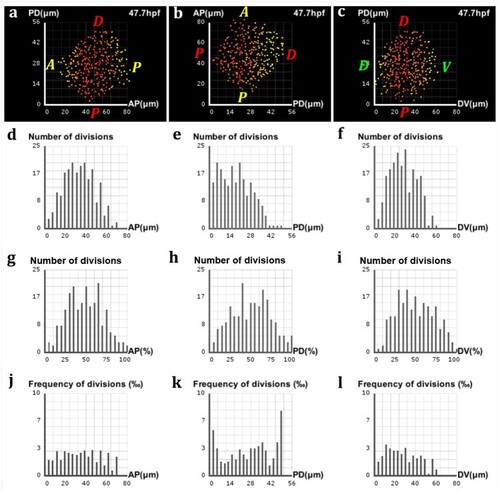
Analysis of proliferation in the zebrafish pectoral fin. (a–c) Frequencies of divisions along the AP, PD and DV axes respectively, highlighted by a yellow-red color gradient coding for differences in proliferation rates across the fin. (a, c) The preponderance of red at the center of the fin shows where the bulk of cell divisions takes place, with only a few of them occurring near the lateral surfaces (yellow). (b) A decreasing gradient of proliferation rates from the proximal pole to the distal tip characterizes the PD axis. (d–f) Marginal distributions of proliferation along the AP, PD and DV axes respectively, expressed in numbers of cells with respect to the absolute distance in μm along the axis. (g–i) Same distributions with respect to the relative distance on the axis. (j–l) Same distributions expressed in proportions of cells with respect to the absolute distance |
|
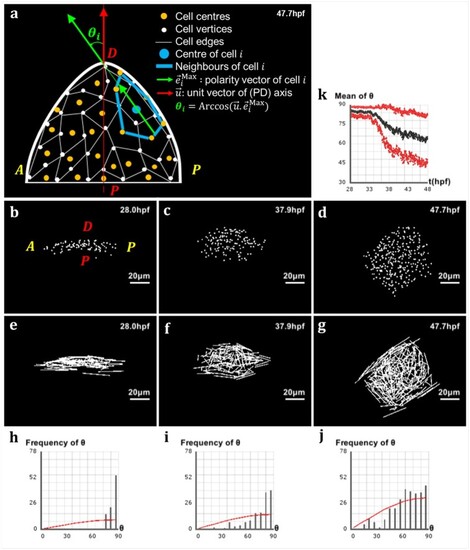
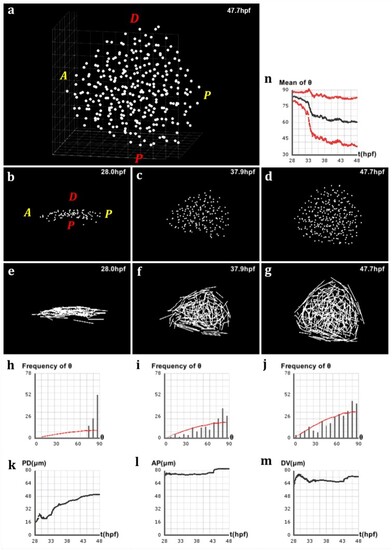
Analysis of directional cell behaviors in the zebrafish pectoral fin. (a) Schematics in 2D of the method used to analyze directional cell behaviors: for each cell i, θi denotes the polarity angle that this cell forms between its elongation axis |
|
Simulation of pectoral fin morphogenesis based on directional cell behaviors. Values of the equation parameters: |