- Title
-
ITPR1 Mutation Contributes to Hemifacial Microsomia Spectrum
- Authors
- Liu, Z., Sun, H., Dai, J., Xue, X., Sun, J., Wang, X.
- Source
- Full text @ Front Genet
|
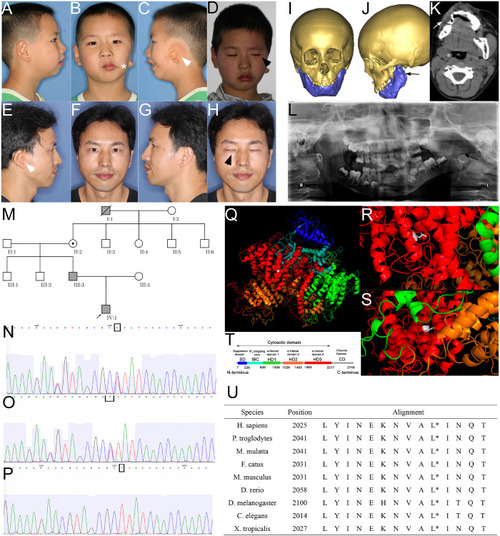
Phenotype and pedigrees of the proband with ITPR1 mutation. |
|
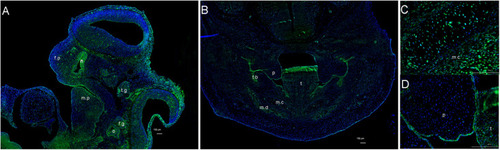
Expression pattern of |
|
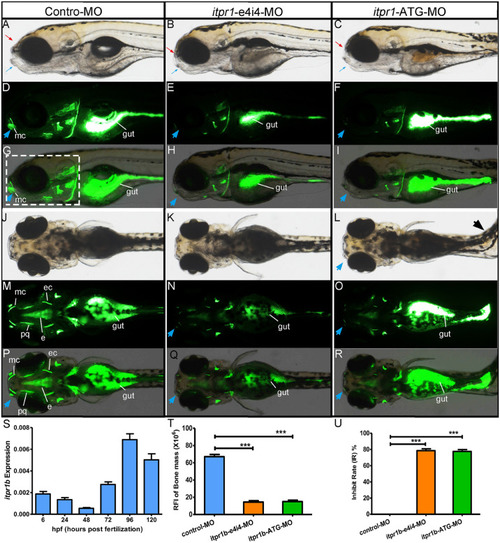
Gross morphology of zebrafish with PHENOTYPE:
|
|
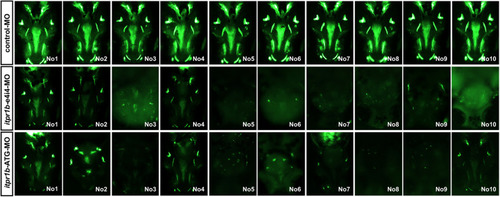
Zebrafish have been picked up to take photos and calculate the craniofacial skeleton mass. PHENOTYPE:
|
|
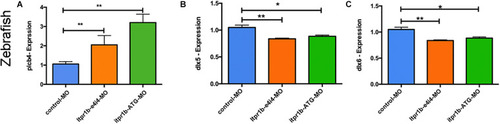
ITPR1 involved in the EDN-PLC-DLX5/6 axis. (A) qRT-PCR analysis of plcb4 expression in zebrafish with itpr1b knockdown was shown, plcb4 transcripts increased in both MO-itpr1-e4i4 and MO-itpr1-ATG morphants. The relative transcript level was calculated as fold change using the 2– ΔΔCt method (ANOVA) (N = 5). (B,C) dlx5 and dlx6 transcripts decreased in both MO-itpr1-e4i4 and MO-itpr1-ATG morphants. (ANOVA, N = 5). |
|
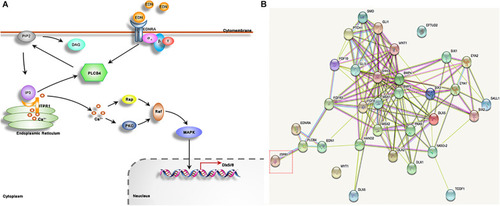
ITPR1 is involved in different pathways. |