- Title
-
Adipose-derived mesenchymal stem cells promote the malignant phenotype of cervical cancer
- Authors
- Castro-Oropeza, R., Vazquez-Santillan, K., Díaz-Gastelum, C., Melendez-Zajgla, J., Zampedri, C., Ferat-Osorio, E., Rodríguez-González, A., Arriaga-Pizano, L., Maldonado, V.
- Source
- Full text @ Sci. Rep.
|
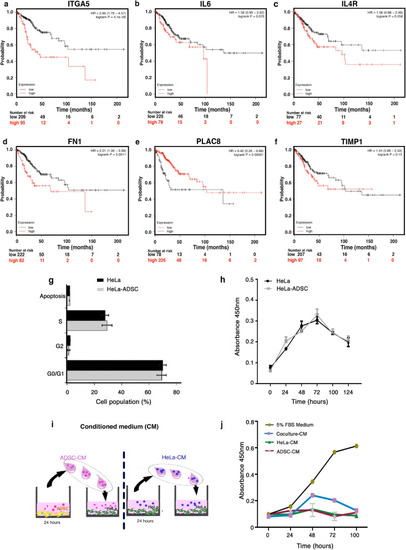
The coculture of HeLa/ADSC induces changes in the transcriptome of HeLa cells. ( |
|
Deregulated genes altered by the presence of ADSC exhibit clinical significance in cervical cancer patients. ( |
|
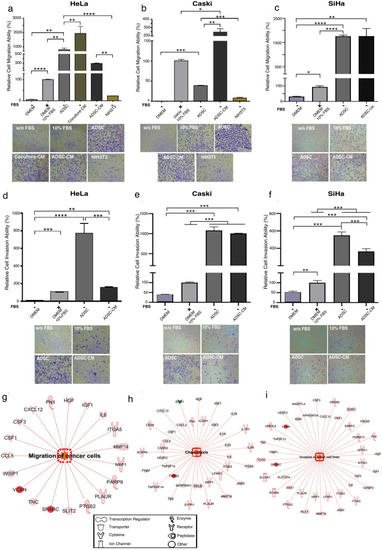
ADSCs influence the migration and invasion of cervical cancer cells. Graph shows the migration ability of HeLa (a), CaSki (b) and SiHa (c) cells cultured without serum and exposed to different chemoattractants including ADSCs, conditioned medium of coculture (Coculture-CM), conditioned medium of ADSCs (ADSC-CM) and NIH3T3 cells. As a control, CC cells were also cultured with DMEM supplemented with 10% FBS or without FBS. The graph shows the relative percentage of the migration capacity of HeLa, CaSki and SiHa cells after 12 h (a–c). The graph represents three biological replicates, error bars are s.d and *p < 0.05. Pictures show a representative image of the migratory CC cells in each condition. (d–f) Figures show the relative percentage of invading HeLa, CaSki and SiHa cells after 12 h of exposure to various chemoattractants. Figures show a representative image of invading cells in each condition. The graph represents three biological replicates, error bars are s.d and *p < 0.05. IPA analysis showing that the main altered transcripts in HeLa cells cultured in presence of ADSCs are involved in migration (g), chemotaxis (h), and invasion (i). The networks show differentially expressed genes regulated by each signaling pathway. The red color indicates the overexpression of the transcripts. |
|
ADSC increases the CSC population, migration and invasion in an in vivo model. ( |
|
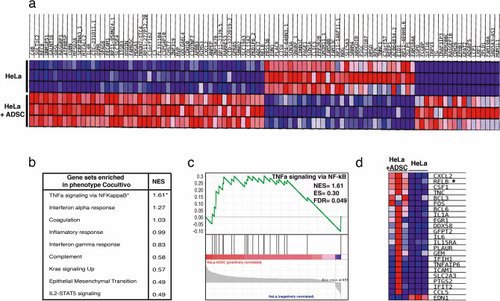
The NF-Kappa B pathway is the most activated pathway during the coculture of HeLa-ADSC. ( |
|
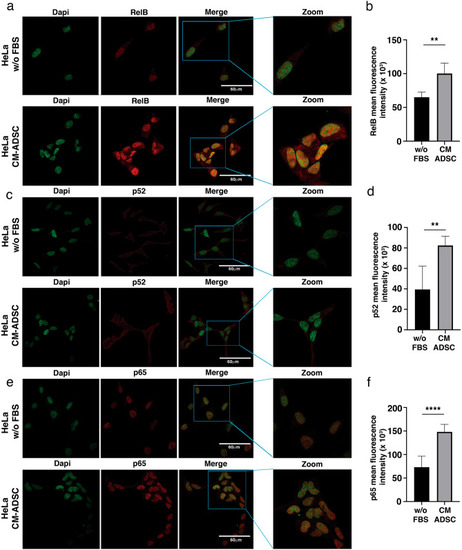
ADSCs induce the expression of the NF-kappa B molecules in CC cells. ( |
|
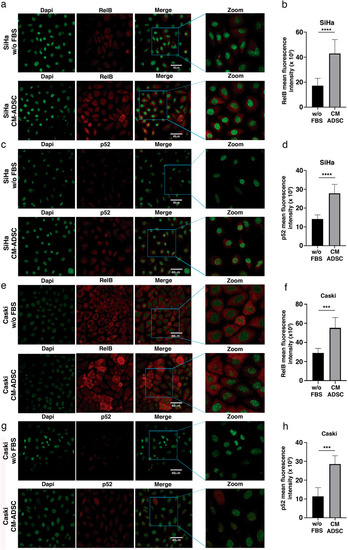
ADSC induces the activation of the NF-kappa B pathway in SiHa and CaSki cells. ( |
|
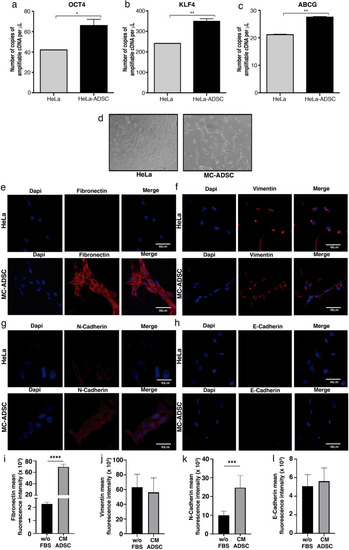
ADSC promotes a stem cell and EMT phenotype in HeLa cells. (a–c) Graphs show the expression level of pluripotency genes evaluated by ddPCR in HeLa control vs HeLa cells cocultured with ADSC. Pluripotency genes include OCT4 (a), KLF4 (b) and ABCG (c). The error bars represent the means ± standard deviation (SD). (d) Schematic representation shows that cervical cancer cells exhibit an EMT-like phenotype induced by the conditioned medium of ADSC. Expression of EMT markers including fibronectin (e), Vimentin (f), N-cadherin (g) and E-cadherin (h) analyzed by immunofluorescence of HeLa cells cultured with or without conditioned ADSC. EMT proteins were stained with Cy3-conjugated secondary antibody and nuclei were stained with 4′-6-Diamidino-2-phenolindole (DAPI). Images were taken in a confocal microscope using an × 40 oil lens. Photographs are representative of three independent experiments. (i–j) To quantify the expression levels of fibronectin (i), vimentin (j), N-cadherin (k) and E-cadherin (l), the intensity of fluorescence was quantified using Image J. Graph represents three biological replicates, error bars are s.d and ****p < 0.0001 and ***p < 0.001. |
|
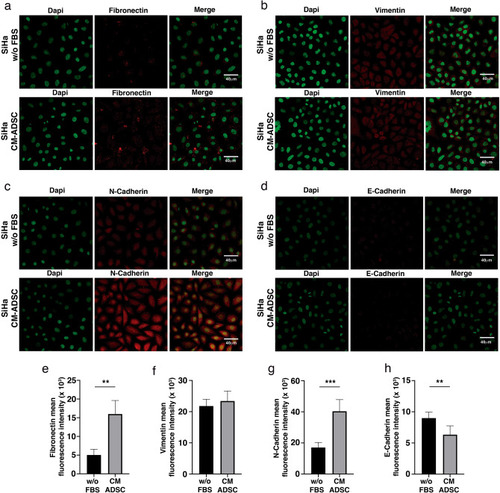
ADSC induce an EMT phenotype in SiHa cells. ( |
|
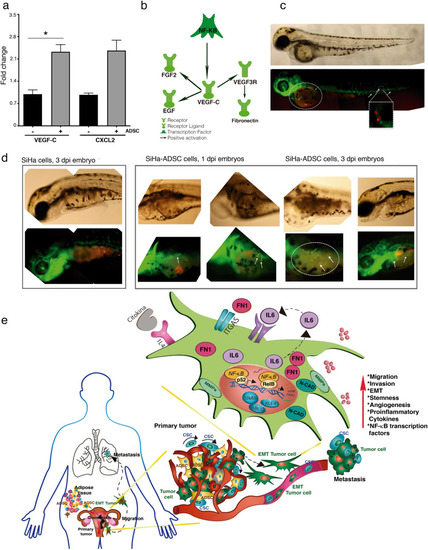
ADSC modulate angiogenesis in CC tumors. (a) Graph shows the fold increase of DE mRNAs involved in angiogenesis obtained from HeLa/ADSC RNAseq data. Angiogenesis-related mRNAs shown are VEGF-C, and CXCL2. (b) Representative network of KPA analysis showing that NF-Kappa B/VEGF-C activates the signaling of angiogenic factors in HeLa cells cocultured in the presence of ADSC. (c) Representative photograph of the infiltration of SiHa cells in the blood vessels of fli1a embryos: EGFP after 12 h post-injection is shown. The SiHa cells are observed in red color and blood vessels in green. (d) Representative figure showing the formation of new blood vessels in the yolk of embryos from day 1 dpi and 3 dpi of SiHa cells vs SiHa-ADSC. (e) The illustration built with the experimental data obtained, indicates that the ADSCs migrate from the adipose tissue to CC tumor and there, they increase the malignant phenotype of the CC cells and promoting metastasis. The ADSC potentiate the malignant phenotype of cervical cancer cells by increasing the non-canonical NF-Kappa B pathway, causing an increase in the expression of chemokines, transcription factors, metalloproteases, integrins, etc., which contribute to the increased migration and invasion capacity of cancer cells. In addition, ADSC activate the EMT process mainly due to the increase of Fibronectin in CC cells. Finally, ADSCs induce the angiogenic potential that plays an important role in tumor progression. It should be mentioned that all these phenotypic changes could be given by the activation of the NF-Kappa B pathway. |