- Title
-
ndufa7 plays a critical role in cardiac hypertrophy
- Authors
- Shi, X., Zhang, Y., Chen, R., Gong, Y., Zhang, M., Guan, R., Rotstein, O.D., Liu, X., Wen, X.Y.
- Source
- Full text @ J. Cell. Mol. Med.
|
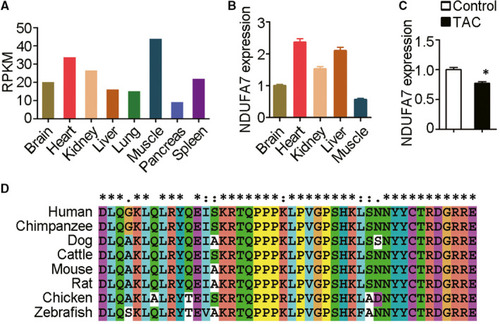
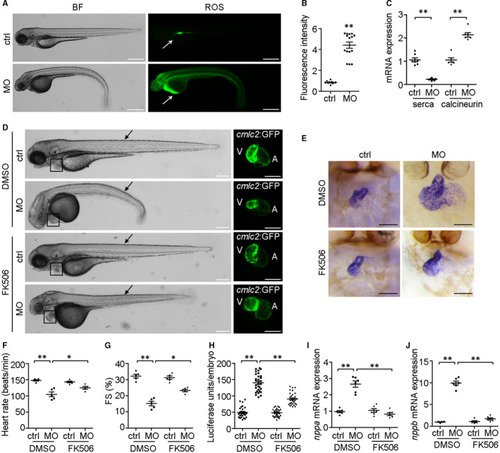
NDUFA7 is involved in cardiac hypertrophy induced by ISO infusion. (A) Tissue expression profiles of NDUFA7 are generated from GTEx database ( |
|
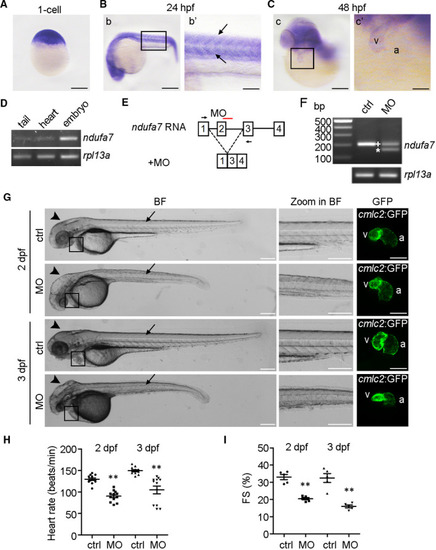
Knockdown of |
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
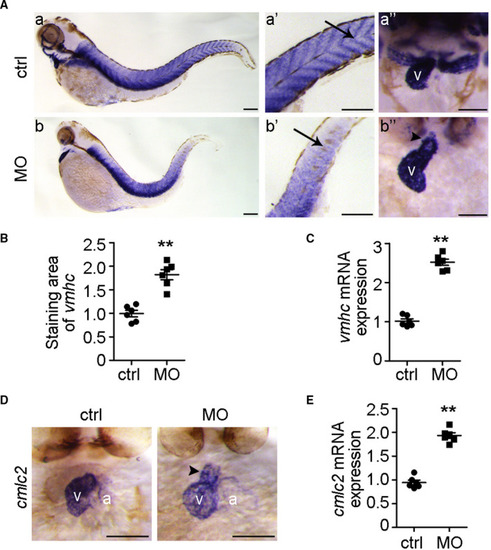
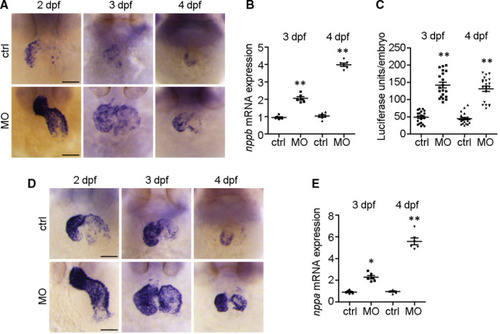
The cardiac hypertrophy biomarkers |
|
Calcium signalling is involved in EXPRESSION / LABELING:
PHENOTYPE:
|
|
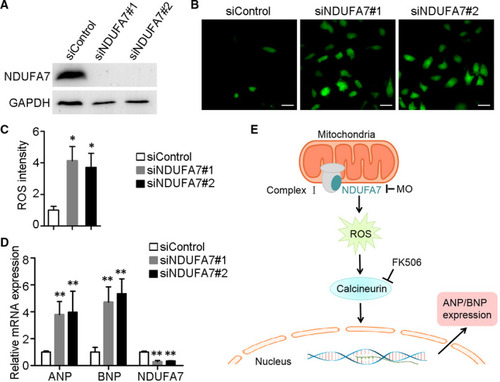
Depletion of NDUFA7 leads to cardiac hypertrophy in H9c2 cells. (A) H9c2 cells were transfected with control or NDUFA7 siRNAs for 72 hours, and expression level of NDUFA7 and GAPDH were examined. (B) H9c2 cells transfected with control or NDUFA7 siRNAs were stained for ROS using 2’,7’‐dichlorofluorescin diacetate, and then visualized under the fluorescence microscope. Scale bar, 50 µm. (C) Experiments were performed as in (B), and the ROS intensity in each group were examined by ImageJ. * |