- Title
-
ADP-dependent glucokinase as a novel onco-target for haematological malignancies
- Authors
- Tandon, A., Birkenhagen, J., Nagalla, D., Kölker, S., Sauer, S.W.
- Source
- Full text @ Sci. Rep.
|
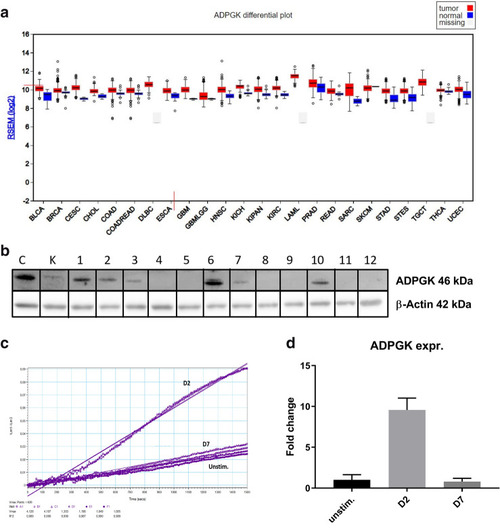
ADPGK activity and expression upon stimulation. ( |
|
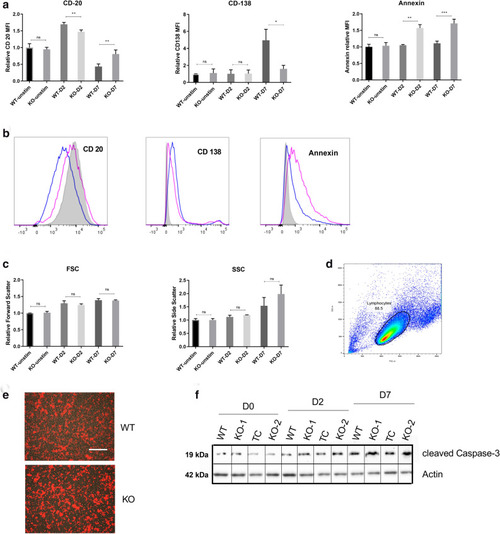
PMA stimulation induced differentiation is hampered in ADPGK KO cells. ( |
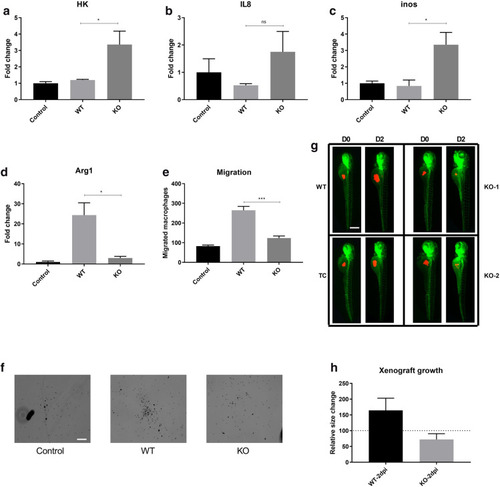
|
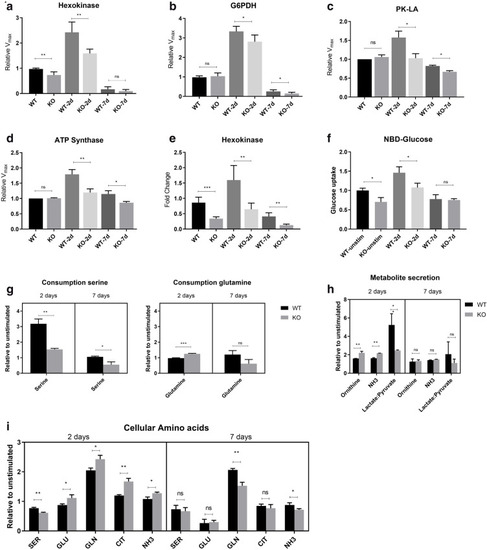
ADPGK KO B-cells are metabolically toxified. (a) |
|
|
|
|
|
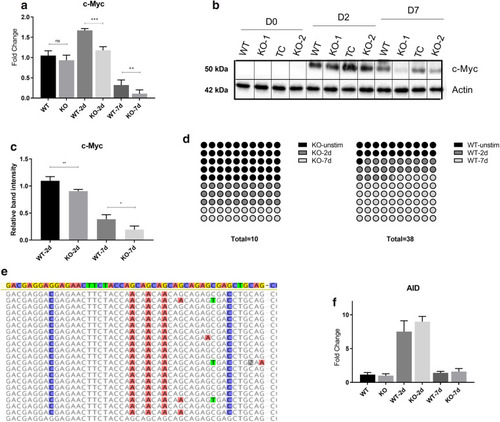
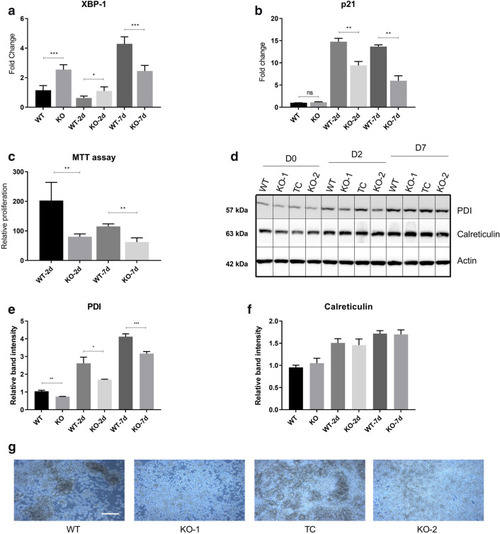
ER stress-based differentiation markers are more expressed in Ramos WT cells. ADPGK KO and Ramos WT cells, treated with PMA and cultured for seven days under standard conditions were analysed via western blot and RT-qPCR at D0, D2 and D7 post stimulation. ( |