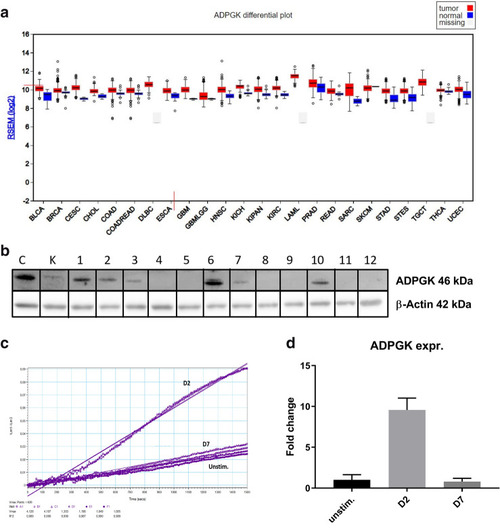
ADPGK activity and expression upon stimulation. (a) Expression data for ADPGK in normal and tumour samples in the TCGA (The Cancer Genome Atlas) FireBrowse expression viewer. Tumour expression- red blocks; Normal tissue expression- blue blocks (b) ADPGK knock-outs were generated via CRISPR/Cas9 technology targeting exon-2 of ADPGK. ADPGK (ADP dependent glucokinase) knock-out selection via western blot using 46 kDa band as reference. β-actin is the loading control. Lanes 1–12 depict 12 analysed clones. Lanes 4 and 8 were selected as KO-1 and KO-2 respectively. Ramos BL (Burkitt's lymphoma) cells were stimulated with 50 ng/ml PMA (phorbol-12 myristate 13-acetate) for a period of seven days and analysed at D2 (two days post stimulation) and D7 (seven days post stimulation). (c) Enzyme kinetics of ADPGK at D2, D7 and unstim. (unstimulated) states. X-axis represents time in seconds and y-axis is difference in absorbance between 340 and 400 nm. (d) Change in ADPGK expression upon stimulation at D2, D7 and unstimulated states using RT-qPCR. Data is representative of three individual experiments. Error bars represent + /- s.e.m. C is positive control and K negative control for ADPGK. TCGA cancer nomenclature is available at https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations.
|

