- Title
-
The extracytoplasmic function sigma factor σVreI is active during infection and contributes to phosphate starvation-induced virulence of Pseudomonas aeruginosa
- Authors
- Otero-Asman, J.R., Quesada, J.M., Jim, K.K., Ocampo-Sosa, A., Civantos, C., Bitter, W., Llamas, M.A.
- Source
- Full text @ Sci. Rep.
|
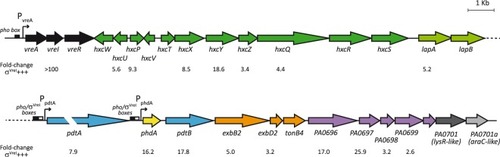
Genetic organization of the σVreI regulon. Transcriptional organization of the |
|
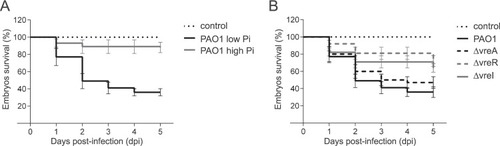
Survival of zebrafish embryos upon infection with |
|
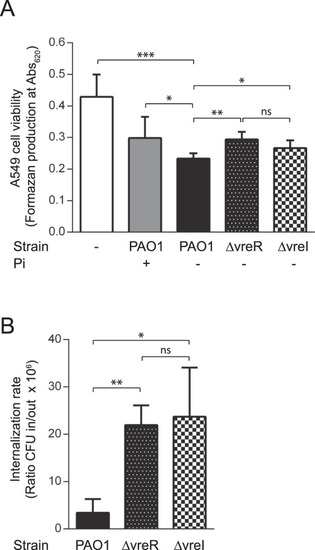
|
|
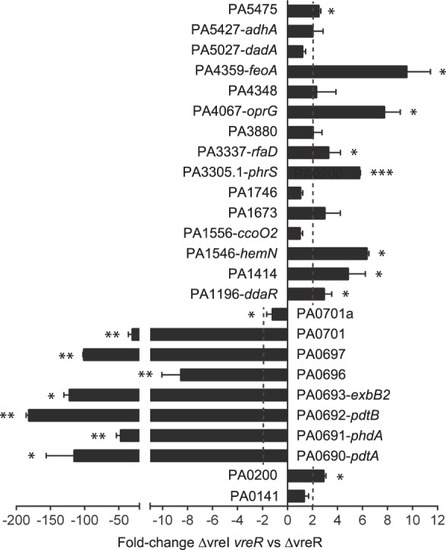
Differential gene expression in the |
|
σVreI activation during PHENOTYPE:
|
|
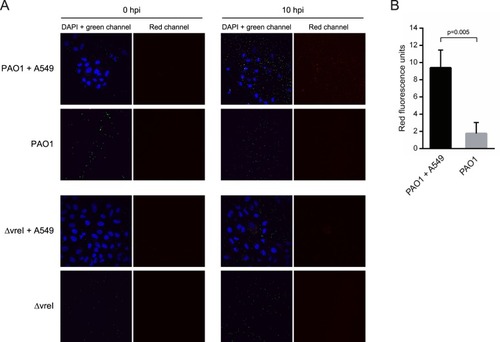
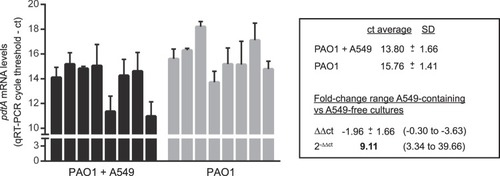
Activation of σVreI upon interaction of |
|
|