- Title
-
Infection and RNA-seq analysis of a zebrafish tlr2 mutant shows a broad function of this toll-like receptor in transcriptional and metabolic control and defense to Mycobacterium marinum infection
- Authors
- Hu, W., Yang, S., Shimada, Y., Münch, M., Marín-Juez, R., Meijer, A.H., Spaink, H.P.
- Source
- Full text @ BMC Genomics
|
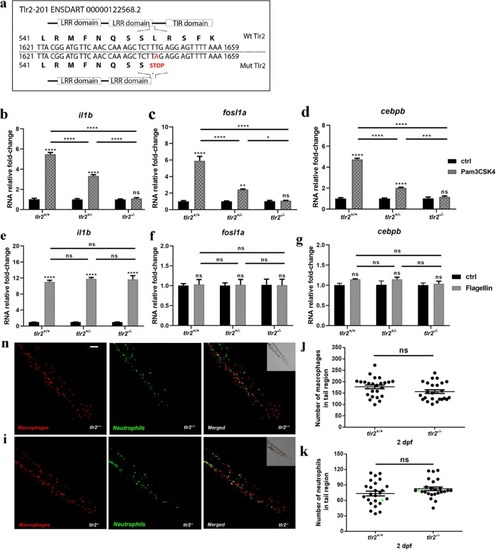
Characterization of the Tlr2 mutant. |
|
Gene expression in the absence of infection between Tlr2 mutants. |
|
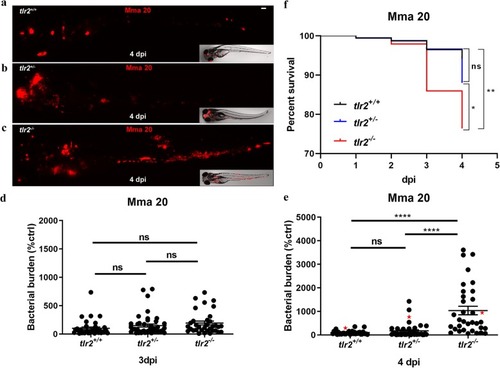
Quantification of bacterial burden and survival after PHENOTYPE:
|
|
Quantification of Mma20 infection phenotype in the PHENOTYPE:
|
|
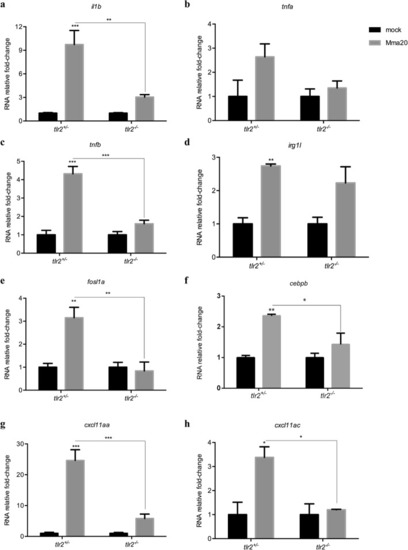
Immune genes expression in |
|
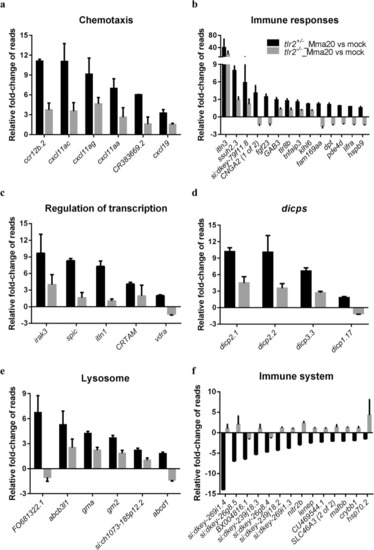
Overview of RNAseq results. |
|
Overview of fold changes of representative genes selected from the gene categories resulting from GO-term analysis. |