- Title
-
Functional Comparison of Human and Zebra Fish FKBP52 Confirms the Importance of the Proline-Rich Loop for Regulation of Steroid Hormone Receptor Activity
- Authors
- Harris, D.C., Garcia, Y.A., Samaniego, C.S., Rowlett, V.W., Ortiz, N.R., Payan, A.N., Maehigashi, T., Cox, M.B.
- Source
- Full text @ Int. J. Mol. Sci.
|
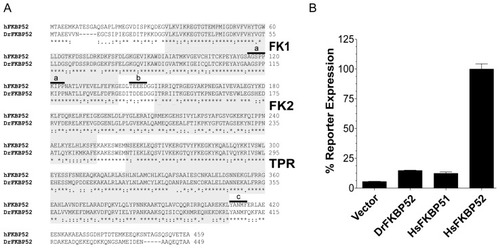
DrFKBP52 contains all known critical domains and residues for function but does not potentiate receptor activity. ( |
|
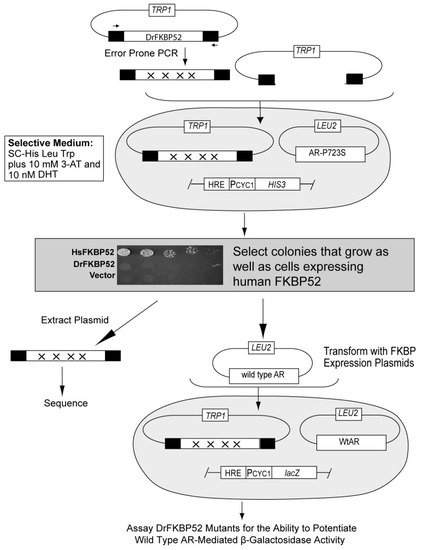
Selection scheme for |
|
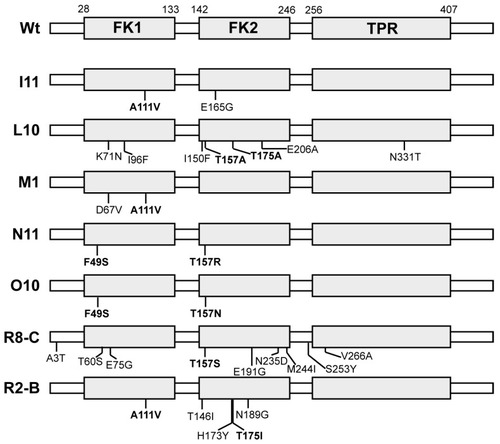
DrFKBP52 gain-of-function mutants isolated. The domain arrangement of wild type DrFKBP52 with amino acid numbering at the domain boundaries is illustrated at the top with identified mutants aligned below. Each mutant is coded based on the independent library from which it was isolated (I to R) and an isolate number. Repeatedly identified mutations of particular interest are shown in bold. |
|
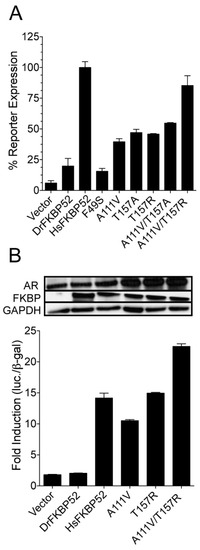
DrFKBP52 mutants potentiate androgen receptor activity. ( |
|
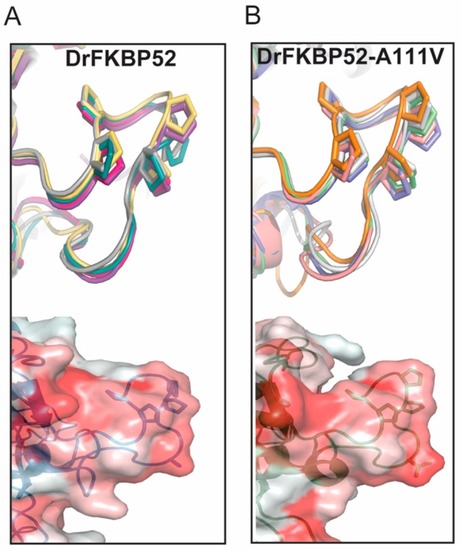
Predictive modeling and hydrophobicity scale of DrFKBP52 and DrFKBP52-A111V. The top structures represent the top five 3D models for each structural query of DrFKBP52 ( |
|
Predicted structural differences in the FK1 proline-rich loop. Homology modeling was used to generate predicted models of DrFKBP52 and DrFKBP52-A111V, in the comparison to human FKBP52 and FKBP51 to determine conformational changes induced by the DrFKBP52-A111V mutation. Crystal structures of HsFKBP51 (PDB ID: 1KT0), HsFKBP52 (PDB ID: 1Q1C), DrFKBP52 (predicted), and DrFKBP52-A111V (predicted) are aligned; the respective FK1 domains are shown in space-filled modeling and colored in hydrophobicity scale (blue = hydrophilic, red = hydrophobic). Note that the only differences between FKBP51 and HsFKBP52 within the loop region are at positions 119 and 124. |