- Title
-
NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish
- Authors
- Qi, J., Dong, Z., Shi, Y., Wang, X., Qin, Y., Wang, Y., Liu, D.
- Source
- Full text @ Cell Res.
|
NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish. (A) Schematic diagram showing the structure of NgAgo-2nls for in vitro transcription and microinjection. (B) Schematic diagram showing guide DNA targeting sites (indicated by arrowhead) in the exons of fabp11a gene. The targeting sequences are highlighted in purple and red. (C) Microscopy analysis of eyes in control and FW-guide DNA 2 and NgAgo-2nls mRNA coinjected embryos, dorsal view, with the head regions enlarged and shown on the right. Red arrowheads indicate normal paired eyes. Blue arrowhead indicates one single eye in FW-guide 2 and NgAgo-2nls mRNA coinjected embryo. Green arrowhead indicates a single fused large eye. (D) Statistical analysis of phenotype efficiency in control (0%); NgAgo-2nls mRNA only (0%); FW-guide 2 only (0%); FW-guide 2 and NgAgo-2nls mRNA (29.6%); FW-guide 2 mismatch 1 and NgAgo-2nls mRNA (0%); RV-guide 2 and NgAgo-2nls mRNA (15.4%); FW-guide 2, RV-guide 2 and NgAgo-2nls mRNA (39.7%); FW-guide 2, NgAgo-2nls mRNA and fabp11a mRNA injected embryos (4.6%). One-Way ANOVA; **** P < 0.0001; * P< 0.05. (E) Relative mRNA levels of zebrafish fabp11a in 30 hpf control; NgAgo-2nls mRNA only; FW-guide 2 only; FW-guide 2 and NgAgo-2nls mRNA; FW-guide 2 mismatch 1 and NgAgo-2nls mRNA; RV-guide 2 and NgAgo-2nls mRNA; FW-guide 2, NgAgo-2nls mRNA and fabp11a mRNA; FW-guide 2, RV-guide 2 and NgAgo-2nls mRNA injected embryos. One-Way ANOVA; **** P < 0.0001. |
|
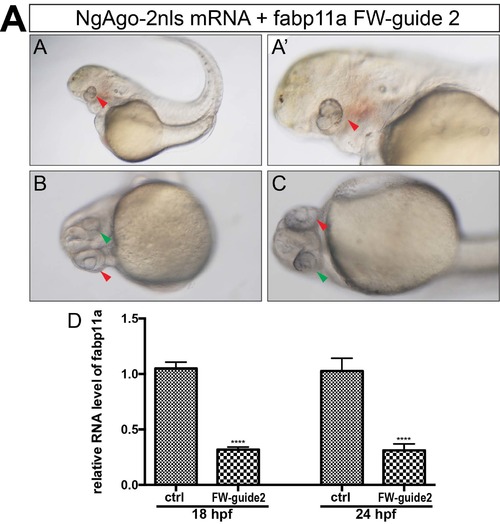
NgAgo based fabp11a gene knockdown caused eye developmental defects in zebrafish. (A, A') Microscopy analysis of eyes infabp11a FW-guide DNA 2 and NgAgo-2nls mRNA coinjected embryos, lateral view. Red arrowheads indicate small developmental abnormal eyes. (B, C) Microscopy analysis of eyes in fabp11a FW-guide DNA 2 and NgAgo-2nls mRNA coinjected embryos, ventral view. Red arrowhead indicates relative normal eye. Green arrowhead indicates small developmental abnormal eye. (D) Relative mRNA levels of zebrafish fabp11a in 18 and 24 hpf control and ta FW-guide DNA 2 and NgAgo-2nls mRNA coinjected embryos. T-test; ****, P<0.0001. |
|
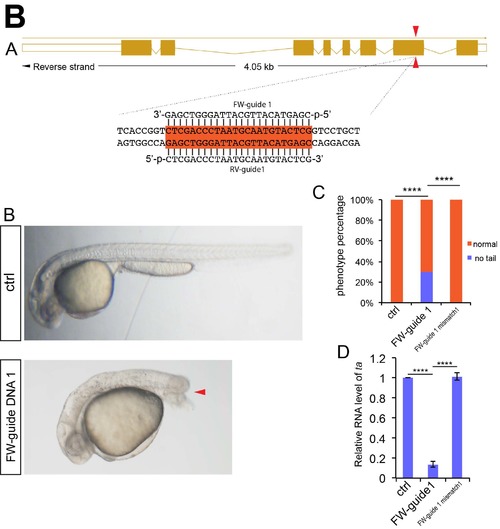
NgAgo based ta gene knockdown caused no tail phenotype in zebrafish. (A) Schematic diagram showing Guide DNA targeting sites on the exon of ta gene. Guide DNA targeting sites are indicated by arrowheads. The targeting sequences are highlighted in orange. (B) Microscopy analysis of tail in control, NgAgo-2nls and ta FW-guide DNA 1 coinjected, and NgAgo-2nls and ta mismatch guide DNA coinjected embryos, lateral view. Red arrowhead indicates disrupted tail. (C) Statistical analysis of phenotype efficiency in control, NgAgo-2nls and ta FW-guide DNA 1 coinjected, and NgAgo-2nls and ta mismatch guide DNA coinjected embryos. χ² test; ****, P<0.0001. (D) Relative mRNA levels of zebrafish ta in 30 hpf control and ta FW-guide DNA 1 and NgAgo-2nls mRNA coinjected embryos. T-test; ****, P<0.0001. |
|
NgAgo based kdrl gene knockdown caused angiogenic defects phenotype in zebrafish. (A) Microscopy analysis of blood vessel in control and NgAgo-2nls and kdrl FW-guide DNA 1 coinjected embryos, lateral view. Green arrowhead indicates intersegmental vessel (ISVs). (B) Relative mRNA levels of zebrafish kdrlin 30 hpf control and kdrl FW-guide DNA 1 and NgAgo-2nls mRNA coinjected embryos. T-test; ****, P<0.0001. (C) Statistical analysis of phenotype efficiency in control and NgAgo-2nls and kdrl FW-guide DNA 1 coinjected embryos. χ² test; ****, P<0.0001. The Tg(kdrl:mCherry) transgenic line was described in our previous work (Jiang Q, Lagos-Quintana M, Liu D, Shi Y, Helker C, Herzog W, et al. miR-30a regulates endothelial tip cell formation and arteriolar branching. Hypertension 2013, 62(3): 592-598.). |
|
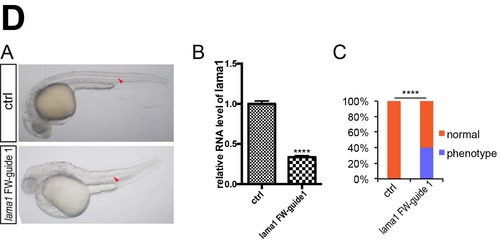
NgAgo based lama1 gene knockdown caused notochord defects phenotype in zebrafish. (A) Microscopy analysis of notochord in control and NgAgo-2nls and lama1 FW-guide DNA 1 coinjected embryos, lateral view. Red arrowhead indicates notochord. (B) Relative mRNA levels of zebrafish lama1 in 30 hpf control and lama1 FW-guide DNA 1 and NgAgo-2nls mRNA coinjected embryos. T-test; ****, P<0.0001. (C) Statistical analysis of phenotype efficiency in control and NgAgo-2nls and lama1 FW-guide DNA 1 coinjected embryos. χ² test; ****, P<0.0001. |
|
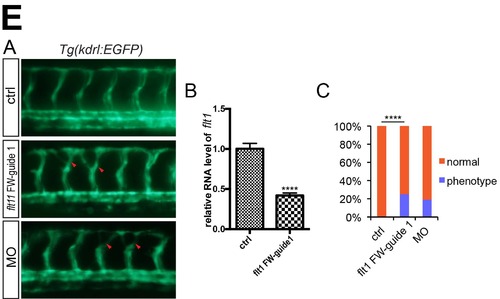
NgAgo based flt1 gene knockdown caused excessive angiogenesis phenotype in zebrafish. (A) Microscopy analysis of blood vessel in control and NgAgo-2nls and flt1 FW-guide DNA 1 coinjected embryos, lateral view. Green arrowhead indicates intersegmental vessel (ISVs). (B) Relative mRNA levels of zebrafish flt1 in 30 hpf control and flt1 FW-guide DNA 1 and NgAgo-2nls mRNA coinjected embryos. T-test; ****, P<0.0001. (C) Statistical analysis of phenotype efficiency in control and NgAgo-2nls and flt1 FW-guide DNA 1 coinjected embryos. χ² test; ****, P<0.0001. The Tg(kdrl:EGFP) transgenic line was described in our previous work (Wang X, Ling CC, Li L, Qin Y, Qi J, Liu X, et al. MicroRNA-10a/10b represses a novel target gene mib1 to regulate angiogenesis. Cardiovascular research 2016, 110(1): 140-150.). |
|
Mutated NgAgo mRNA coinjected with fabp11a FW-gDNA2 and ta FW-gDNA1 respectively caused eye phenotype and no tail phenotype. (A) Microscopy analysis of eyes in ctrl, fabp11a FW-guide DNA 2 and NgAgo-D663A mRNA coinjected, fabp11a FW-guide DNA 2 and NgAgo-D738A mRNA coinjected, and fabp11a FW-guide DNA 2 and NgAgo-D663A-D738A mRNA coinjected embryos. (B) Microscopy analysis of eyes in ctrl, ta FW-guide DNA 1 and NgAgo-D663A mRNA coinjected, ta FW-guide DNA 1 and NgAgo-D738A mRNA coinjected, and ta FW-guide DNA 1 and NgAgo-D663A-D738A mRNA coinjected embryos. |
|
Whole mount in situ hybridization analysis of zebrafish embryos using antisense fabp11a probe. (A) 11.5 hpf, lateral view. Red arrowhead indicates eye field. (A'). 11.5 hpf, coronal view. (B) 12 hpf, lateral view. Red arrowhead indicates eye field. (B'). 12 hpf, coronal view. (C) 14 hpf, lateral view. Red arrowhead indicates eye field. (C'). 14 hpf, coronal view. (D) 16 hpf, lateral view. Red arrowhead indicates eye field. (D'). 16 hpf, coronal view. (E) 18 hpf, lateral view. Red arrowhead indicates eye field. (E'). 18 hpf, coronal view. (F) 18.5 hpf, lateral view. Red arrowhead indicates eye field. (F'). 18.5 hpf, coronal view. (G) 19 hpf, lateral view. Red arrowhead indicates eye field. (G'). 19 hpf, coronal view. (H) 20 hpf, lateral view. Red arrowhead indicates eye field. (H'). 20 hpf, coronal view. EXPRESSION / LABELING:
|
|
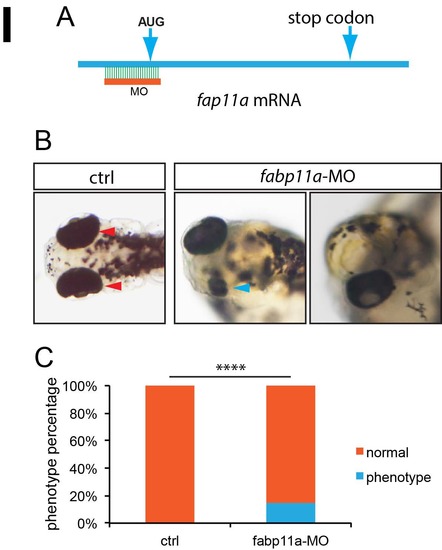
Fabp11a gene knockdown by morpholino resulted in eye developmental defects in zebrafish. (A) Diagram of fabp11a translation blocking morpholino design. (B) Microscopy analysis of eyes in control and fabp11a morphants, dorsal view. Red arrowheads indicate normal paired eyes. Blue arrowhead indicates normal small eyes. (C) Statistical analysis of phenotype efficiency in control and fabp11a MO injected embryos. χ² test; ****, P<0.0001. PHENOTYPE:
|
|
Fabp11a gene knockout using CRISPR/Cas9 system caused eye developmental defects in zebrafish. (A) Schematic diagram showing Guide RNA targeting sites on the exons of fabp11a gene. Guide DNA targeting sites are indicated by arrowheads. The targeting sequences are highlighted in orange and purple. (B) Graphical view of Fabp11a protein domain. (C) Sanger sequencing analysis of PCR fragments amplified from gRNA1 and gRNA2 target regions in gRNA1/gRNA2 and cas9 mRNA coinjected embryos. (D) Mutation pattern of gRNA1/gRNA2 and cas9 mRNA coinjected embryos. Numbers in the brackets show the number of nucleotides were deleted (-) or inserted (+). Inserted nucleotide is in red. WT, wild type. (E) Microscopy analysis of eyes in control and gRNA1-cas9 mRNA coinjected embryos, dorsal view. Red arrowheads indicate normal paired eyes. Blue arrowhead indicates no eye in gRNA1-cas9 mRNA coinjected embryo. Green arrowhead indicates single fused big eyes, dorsal view. Purple arrowhead indicates single eye, dorsal view. (F) Statistical analysis of phenotype efficiency in control and gRNA1-cas9 mRNA coinjected embryos. χ² test; ****, P<0.0001. (G) Microscopy analysis of eyes in control and fabp11a-/- embryos. Arrowheads indicate eyes. PHENOTYPE:
|