- Title
-
The BH3-Only SNARE BNip1 Mediates Photoreceptor Apoptosis in Response to Vesicular Fusion Defects
- Authors
- Nishiwaki, Y., Yoshizawa, A., Kojima, Y., Oguri, E., Nakamura, S., Suzuki, S., Yuasa-Kawada, J., Kinoshita-Kawada, M., Mochizuki, T., and Masai, I.
- Source
- Full text @ Dev. Cell
|
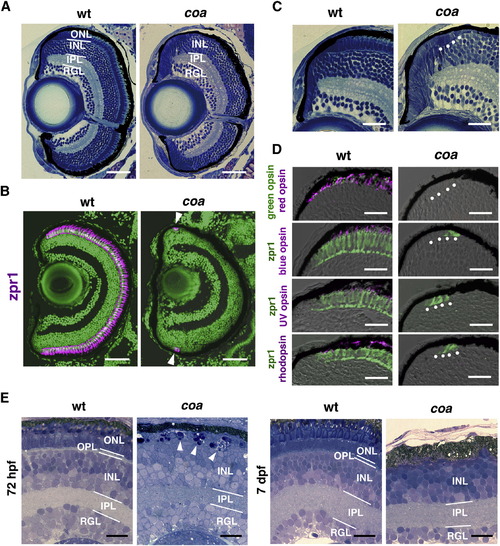
Photoreceptors Undergo Apoptosis in the Zebrafish coa Mutant (A) Plastic sections of WT and coa mutant retinas at 6 dpf. ONL is absent in the coa mutant. (B) Zpr1 antibody labeling of 6 dpf WT and coa mutant retinas. All nuclei are counterstained with SYTOX Green (green). Arrowheads indicate zpr1 expression (magenta) in the CMZ of the coa mutant. (C) Plastic sections of CMZ of 6 dpf WT and coa mutant retinas. White dots indicate morphologically distinct ONL in the coa mutant. (D) Labeling of 6 dpf WT and coa mutant retinas with anti-red, blue, UV opsins, or rhodopsin antibodies (magenta); anti-green opsin antibody (green); and zpr1 (green) antibody. (E) Semithin sections of WT and coa mutant retinas at 72 hpf and 7 dpf. At 72 hpf, pyknotic nuclei were observed in the ONL in the coa mutant (arrowheads). At 7 dpf, ONL was absent in the coa mutant. INL, inner nuclear layer; IPL, inner plexiform layer; OPL, outer plexiform layer; RGL, retinal ganglion cell layer. Scale bars, 50 μm (A and B) and 20 μm (C–E). See also Figure S1. EXPRESSION / LABELING:
PHENOTYPE:
|
|
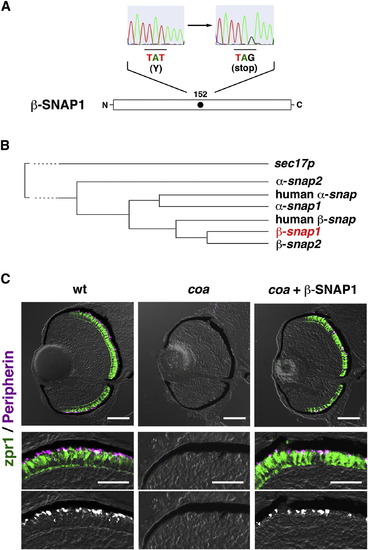
The coa Mutant Gene Encodes β-SNAP1(A) A non-sense mutation occurs at 152Y in β-snap1 gene in the coa mutant.(B) There are four zebrafish snap genes: two α-snap and two β-snap genes. β-SNAP1 has 90.8% amino acid identity to β-SNAP2, whereas β-SNAP1 has 78% and 73% amino acid identity to α-SNAP1 and α-SNAP2, respectively. In accordance with the evolution tree, zebrafish α-snap2 gene is positioned outside the human α-snap gene. Zebrafish β-snap genes seem to be duplicated after separation from human β-snap gene. sec17p is a yeast α-SNAP homologous gene. (C) Labeling of 84 hpf WT, coa mutant retinas, and coa mutant retinas expressing β-SNAP1 with zpr1 antibody (green) and GFP-tagged peripherin (magenta). Middle panels show higher magnification of the outer photoreceptor layer. Lower panels show the magenta channel. Scale bars, 50 μm (upper panels) and 10 μm (middle panels). See also Figure S2. EXPRESSION / LABELING:
|
|
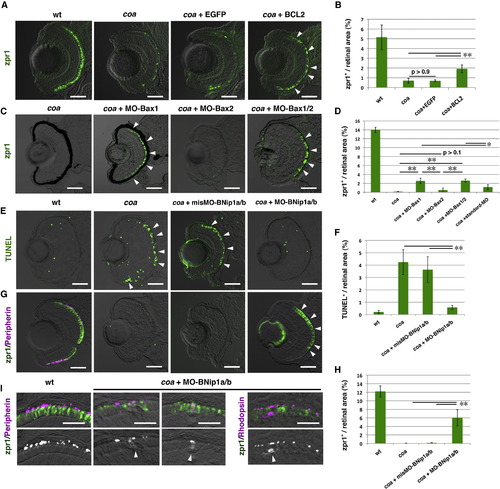
Photoreceptor Degeneration in the coa Mutant Depends on BNip1 (A) Zpr1 labeling (green) of 84 hpf WT, coa mutants, coa mutants injected with egfp mRNA, and bcl2 mRNA. Arrowheads indicate rescued photoreceptors. (B) Percentage of zpr1-positive areas relative to the total retinal area in the experiments shown in (A). Green and black bars indicate the mean and SD, respectively. **p < 0.01. (C) Zpr1 labeling (green) of 84 hpf coa mutant and coa mutants injected with MO-Bax1, MO-Bax2, or a mixture of MO-Bax1 and MO-Bax2. Arrowheads indicate rescued photoreceptors. (D) Percentage of zpr1-positive areas relative to the total retinal area in the experiments shown in (C) as well as in a coa mutant injected with the standard MO. Green and black bars indicate the mean and SD, respectively. **p < 0.01 and *p < 0.05. (E) TUNEL staining (green) of 84 hpf WT, coa mutant, coa mutant retinas injected with misMO-BNip1a/b, and a mixture of MO-BNip1a and MO-BNip1b. Arrowheads indicate apoptosis in the ONL. (F) Percentage of TUNEL-positive areas relative to the total retinal area in the experiments shown in (E). Green and black bars indicate the mean and SD, respectively. **p < 0.01. (G) Zpr1 labeling (green) and GFP-tagged peripherin (magenta) in 84 hpf WT, coa mutant, coa mutant retinas injected with misMO-BNip1a/b or a mixture of MO-BNip1a and MO-BNip1b. Arrowheads indicate rescued photoreceptors. Nonspecific zpr1 signal was observed in the lens. (H) Percentage of zpr1-positive areas relative to the total retinal area in the experiments shown in (G). Green and black bars indicate the mean and SD, respectively. **p < 0.01. (I) Zpr1 labeling (green) and GFP-tagged peripherin or rhodopsin (magenta) in the photoreceptor cell layer in WT and coa mutants injected with MO-BNip1a and MO-BNip1b. Lower panels show the magenta channel. Arrowheads indicate the failure of transport to the OS.The numbers of retinal sections used in the experiments shown in (B), (D), (F), and (H) and p values for the t test are shown in Table S2. Scale bars, 50 μm (A, C, E, and G) and 10 μm (I). See also Figures S3 and S4. EXPRESSION / LABELING:
PHENOTYPE:
|
|
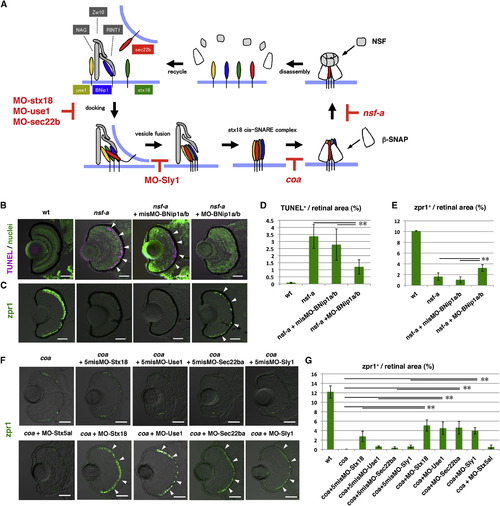
The Syntaxin-18 cis-SNARE Complex Is Required for BNip1-Dependent Apoptosis(A) Molecular mechanism underlying vesicular fusion to the ER. Interactions among syntaxin-18 (green), BNip1 (blue), Use1 (yellow), and Sec22b (red) initiate the fusion of transport vesicles to the ER membrane. Three accessory proteins, Zw10, NAG, and RINT1, play a role in the tethering of transport vesicles. After membrane fusion occurs, β-SNAP binds to the cis-SNARE complex and recruits NSF. NSF promotes disassembly of the cis-SNARE complex. In the absence of β-SNAP or NSF activity, the syntaxin-18 cis-SNARE complex accumulates. MOs against syntaxin-18, Use1, Sec22b, and Sly1 inhibit the formation of the syntaxin-18 cis-SNARE complex, even in the absence of β-SNAP or NSF activity. (B) TUNEL staining (magenta) of 72 hpf WT, nsf-a mutant, and nsf-a mutant retinas injected with misMO-BNip1a/b, and a mixture of MO-BNip1a and MO-BNip1b. Nuclei were counterstained with SYTOX Green (green). Arrowheads indicate apoptosis in the ONL. (C) Zpr1 labeling (green) of 84 hpf WT, nsf-a mutant, nsf-a mutant retinas injected with misMO-BNip1a/b, and a mixture of MO-BNip1a and MO-BNip1b. Arrowheads indicate rescued photoreceptors. (D and E) Percentage of TUNEL-positive (D) and zpr1-positive areas (E) relative to the total retinal area in the experiments shown in (B) and (C), respectively. Green and black bars indicate the means and SDs, respectively. **p < 0.01. (F) Zpr1 labeling (green) of coa mutant and coa mutants injected with MO-syntaxin-18, MO-Use1, MO-Sec22ba, MO-Sly1, their 5misMOs, and MO-syntaxin 5al. Arrowheads indicate rescued photoreceptors. (G) Percentage of zpr1-positive area relative to the total retinal area in the experiments shown in (F). Green and black bars indicate the means and SDs, respectively (**p < 0.01). The numbers of retinal sections used in the experiments shown in (D), (E), and (G) and p values for the t test are shown in Table S2. Scale bars, 50 μm. See also Figure S5. EXPRESSION / LABELING:
PHENOTYPE:
|
|
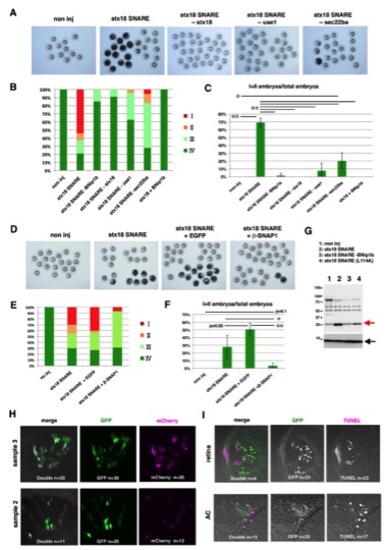
Concurrent Expression of Syntaxin-18 SNARE Components Induces Bax-Dependent Apoptosis in a BNip1-BH3 Domain-Dependent Manner(A) Antiactivated caspase 3 antibody labeling of 7 hpf embryos injected with different RNA combinations of syntaxin-18 SNARE components: noninjection, syntaxin-18 SNARE mixture, syntaxin-18 SNARE mixture lacking BNip1b, syntaxin-18 SNARE mixture replaced with BNip1b (L114A), and syntaxin-18 SNARE mixture plus a mixture of MO-Bax1 and MO-Bax2. Cells with active caspase 3 were increased only in embryos injected with a mixture of mRNAs for all four syntaxin-18 SNARE components. (B) Top panels indicate 8 hpf embryos injected with the same RNA combinations shown in (A). The bottom panel shows a classification of the morphological defects of 8-hpf-injected embryos as embryonic cell death (class I), blown-up blastoderm during gastrulation (class II), nonuniform thickness of the blastoderm (class III), and normal morphology (class IV). (C) Percentages of 8 hpf embryos classified as classes I–IV in the injection experiments shown in (B) as well as the injection experiments for the syntaxin-18 SNARE mixture plus either MO-Bax1 or MO-Bax2. (D) Percentage of class I/II embryos. Green and black bars indicate the mean and SD, respectively. The numbers of injection experiments and t test p values are shown in Table S2 . *p < 0.05, **p < 0.01. (E) TUNEL staining of WT embryos expressing a mixture of all syntaxin-18 SNARE components and EGFP or only EGFP under the control of the atoh7 retinal enhancer. In embryos expressing a mixture of all syntaxin-18 SNARE components and EGFP, TUNEL-positive cells (magenta) are observed in the GFP-positive (green) retinal region, especially retinal ganglion cells (RGC; arrowheads), and anterior commissural (AC, arrowheads) and olfactory (olf, arrow) neurons. (F) The density of apoptotic cells (number/100 μm2) in retinas injected with DNA constructs encoding a mixture of syntaxin-18 SNARE components and EGFP or only EGFP. The number of examined retinas was n = 4 for each. Green and black bars indicate the mean and SD; **p < 0.01. Scale bars, 200 μm (A), 600 μm (B), and 50 μm (E). See also Figure S6. |
|
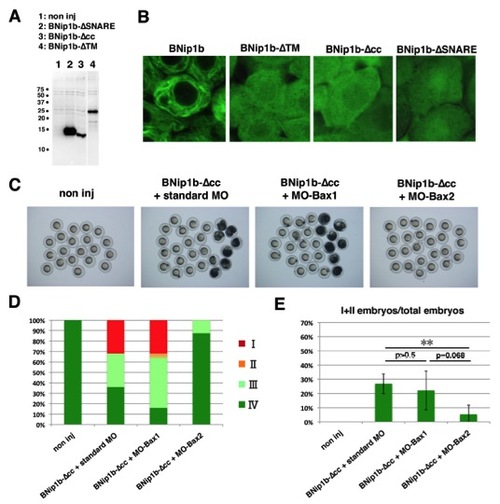
The N-Terminal Coiled-Coil Domain Suppresses BNip1 Proapoptotic Activity(A) BNip1b has four distinct domains: a coiled-coil domain, a BH3 domain, a SNARE domain, and a TM domain. Configuration of deletion constructs (BNip1b-ΔTM, BNip1b-ΔSNARE, BNip1b-Δcc, BNip1b-Δcc-BH3(L114A), and BNip1b-Δcc-ΔBH3). (B) Activation of caspase 3 at 7 hpf (upper rows) and morphologies at 8 hpf (lower rows) in embryos injected with mRNA encoding BNip1b, BNip1b-ΔTM, BNip1b-ΔSNARE, BNip1b-Δcc, and BNip1b-Δcc-ΔBH3. Severe embryonic death and morphological defects are observed only in BNip1Δcc-expressing embryos. (C) Percentages of 8 hpf embryos classified as classes I–IV in the injection experiments shown in (B) (lower panels), as well as BNip1b-Δcc-BH3(L114A). (D) Percentage of class I/II embryos. Green and black bars indicate the means and SDs, respectively. The numbers of injection experiments and t test p values are shown in Table S2. **p < 0.01. Scale bars, 200 μm (B, upper panels) and 600 μm (B, lower panels). See also Figure S7. |
|
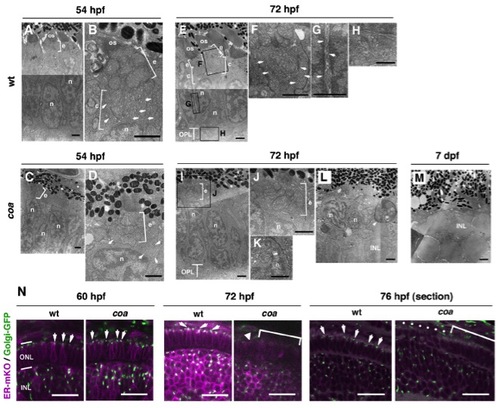
Amounts of OS and intracellular cisternae are reduced in coa mutant photoreceptors, related to Figure 1 |
|
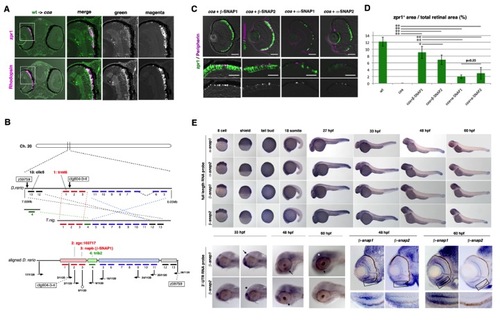
Cloning of the coa mutant gene, related to Figure 2 |
|
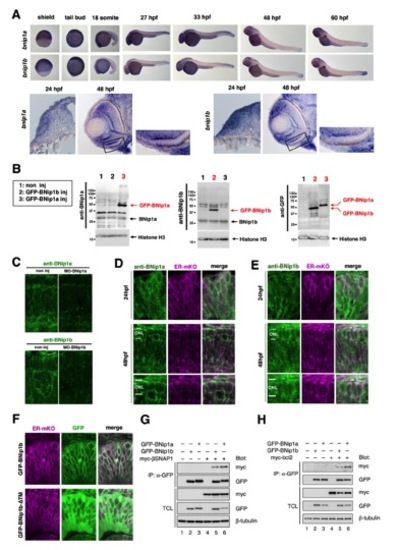
Expression of zebrafish BNip1 mRNA, subcellular localization of zebrafish BNip1 protein, and the in vitro interaction between zebrafish BNip1, !-SNAP and Bcl2 proteins, related to Figure 3 |
|
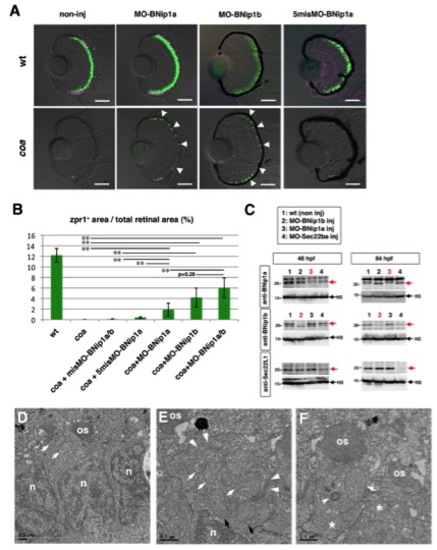
MO-BNip1 rescues photoreceptors but not defects in OS and intracellular cisternae formation, related to Figure 3 |
|
Injection experiments of a mixture of mRNAs and DNA constructs for syntaxin 18 SNARE components, related to Figure 5 |
|
Injection experiments of a mixture of mRNAs for BNip1 deletion mutants, related to Figure 6 |
Reprinted from Developmental Cell, 25(4), Nishiwaki, Y., Yoshizawa, A., Kojima, Y., Oguri, E., Nakamura, S., Suzuki, S., Yuasa-Kawada, J., Kinoshita-Kawada, M., Mochizuki, T., and Masai, I., The BH3-Only SNARE BNip1 Mediates Photoreceptor Apoptosis in Response to Vesicular Fusion Defects, 374-387, Copyright (2013) with permission from Elsevier. Full text @ Dev. Cell