- Title
-
The transcription factor Vox represses endoderm development by interacting with Casanova and Pou2
- Authors
- Zhao, J., Lambert, G., Meijer, A.H., and Rosa, F.M.
- Source
- Full text @ Development
|
Structure, endoderm-inducing activity and subcellular localization of wild-type cas and a deletion variant. (A) D-Cas contains a deletion of the conserved EFDQYLN peptide located in the C-terminal region (red in the wild type) that is likely to act as an activation domain. cas and D-cas RNAs were used for injection. (B-F) Zebrafish embryos were left uninjected (B) or injected with the RNA indicated bottom right and probed by in situ hybridization for sox17 (B-D) or by immunohistochemistry with anti-myc antibody (E,F). The inset in F indicates the position of myc-positive nuclei (arrows). (G) Immunoprecipitation (IP) for myc-D-Cas and myc-Cas. The myc-Cas and myc-D-Cas proteins are detected at 45 kDa. |
|
In vitro binding assay between Cas, Pou2 and Vox. (A-C,E-G) Co-IP experiments. The RNAs indicated below each panel were translated in vitro in the presence (untagged proteins) or absence (myc- or HA-tagged proteins) of radiolabeled methionine, except for C, where tagged protein was also radiolabeled, then processed for IP with the relevant antibody and resolved by SDS-PAGE. The expected position of the tested proteins is indicated on the left of each panel. Note that Vox and Pou2 artifactually migrate as a doublet in A,B,G. (D) Structure of the vox RNA variant open reading frames used for translation, illustrating the deletions in the ΔN-Vox and ΔC-Vox variants. |
|
Effect of cas, vox and pou2 overexpression on endoderm formation. (A-E,G-J) Zebrafish embryos were either left uninjected (A,G) or injected with the RNA indicated bottom right (B-E,H-J), then processed for in situ hybridization with a sox17 probe. (F,K) qRT-PCR analysis. Embryos were injected with the indicated RNA(s). RNA was then extracted and processed for qRT-PCR analysis of sox17. Levels relative to uninjected controls (WT, set at 1) are plotted. For statistical analyses, samples cas+vox or cas+ved or cas+vent are compared with cas (F) or cas+pou2+vox with cas+pou2 (K); ***P<0.005; error bars indicate s.d. |
|
Structure-function analyses of effect of vox activity on Cas function. (A-E,G-J) Zebrafish embryos were either left uninjected (A,G) or injected with the RNA indicated bottom right (B-E,H-J) and allowed to develop until 80% epiboly, then processed for in situ hybridization with sox17 probe. (F,K) qRT-PCR analysis as in Fig. 3. For statistical analyses, samples cas+vox or casΔN-vox or cas+ΔC-vox are compared with cas RNA-injected samples (F) and vox, EnR-ΔN-vox, VP16-ΔN-vox with uninjected samples (K); ***P<0.005, **P<0.05; error bars indicate s.d. EXPRESSION / LABELING:
|
|
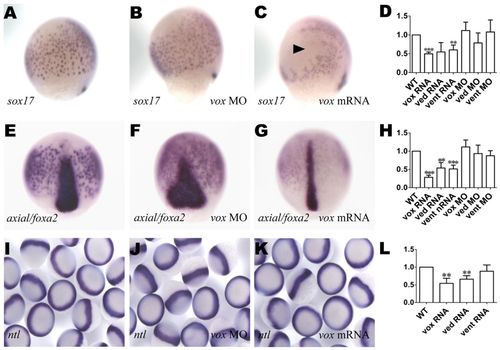
vox is required for endogenous endoderm development. (A-C,E-G,I-K) Zebrafish embryos were either left uninjected (A,E,I) or injected with the RNA or MO indicated bottom right, then processed for in situ hybridization with sox17 (A-C; lateral view with dorsal to the right), axial (E-G; dorsal view) or ntl (I-K) probe. Arrowhead in C indicates a region depleted of sox17-expressing cells. (D,H,L) qRT-PCR analysis (as in Fig. 3) for sox17 (D), axial (H) and gata5 (L). ***P<0.005, **P<0.05, versus uninjected controls; error bars indicate s.d. EXPRESSION / LABELING:
|
|
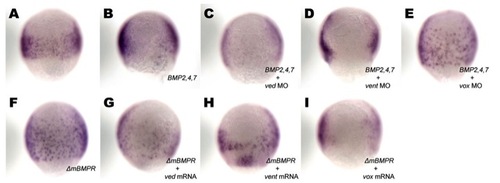
Epistatic analysis of vox and BMPs during endoderm specification. Zebrafish embryos were either left uninjected (A) or injected with the RNA/MO combination indicated bottom right (B-I), allowed to develop until the 75% epiboly stage, then processed for in situ hybridization with sox17. Ventral views are presented. EXPRESSION / LABELING:
|
|
vox represses endodermal fates. Cells were grafted from donor sphere stage zebrafish embryos expressing either Tar*+GFP (A-D) or Tar*+vox+GFP RNAs (E-J) into host uninjected embryos from the same stage, which were allowed to develop and then examined by a combination of epifluorescence and brightfield microscopy at 30 hpf, except in F, which presents a 3-dpf embryo. Lateral views with anterior to the left, except for I which is a dorsal view. (A-D) The main fates induced by Tar* (a constitutively active form of taram-a) expression: pharyngeal endoderm (arrowheads in A,B, where the arrowhead in B points to the forming endodermal pharyngeal pouch), intestine (C, the inset from A; arrowheads points to two cells within the intestine, the contour of which is delineated by dotted lines) and the mesendodermal hatching gland (arrowheads in D). (E-J) The fates induced in Tar*+vox RNA-expressing cells: (E) trunk muscle cells (arrowheads) within the somites (delineated by dotted lines); (F) mesenchymal cells (surrounding the notochord, black arrowheads) and neural tube (white arrowheads); (G) hindbrain cells with their characteristic elongated shape; (H,I) posterior neural tube (arrowheads in H), I is a magnified dorsal view from the inset in H; (J) otic vesicle (black arrowheads) and epidermis (white arrowheads). e, eye; mhb, mid-hindbrain boundary; not, notochord; nt, neural tube; yb, yolk ball; yt, yolk tube. |