- Title
-
Fast Homozygosity Mapping and Identification of a Zebrafish ENU-Induced Mutation by Whole-Genome Sequencing
- Authors
- Voz, M.L., Coppieters, W., Manfroid, I., Baudhuin, A., Von Berg, V., Charlier, C., Meyer, D., Driever, W., Martial, J.A., and Peers, B.
- Source
- Full text @ PLoS One
|
The m1045 mutant exhibits hypoplasia of exocrine pancreas, eyes and branchial arches. (A,B) : WISH using a trypsin probe of unaffected siblings (A) and m1045 mutant embryos (B) at 3.5 days post fertilization (dpf). (C–D) Dorsal view of fluorescent 3.5 dpf unaffected siblings (C) and m1045 mutants (D) in the transgenic ptf1:GFP background. (E,F) Haematoxylin/eosin staining of transverse sections of 4 dpf unaffected siblings (C) and m1045 mutants (D). Alcian blue staining of the cartilage of 3.5 dpf unaffected siblings (C) and m1045 mutants (D). A–D, G–H : views are dorsal; anterior part to the left. p: pancreas. EXPRESSION / LABELING:
PHENOTYPE:
|
|
The m1045 mutant displays a complete loss of cell proliferation in all tissues at 4 dpf. 1-hour Edu incorporation (95–96 hfp) of unaffected siblings (A,C) and m1045 mutant embryos (B,D) in the transgenic ptf1:GFP,ins:dsred background. Confocal projections of 4 dpf embryos of the pancreatic region (A,B) (lateral view) and of the head region (ventral view). Scale bars A,B 50 μm; C,D 100 μm. CMZ: ciliary marginal zone. PHENOTYPE:
|
|
Scheme depicting the strategy used to map and identify the m1045 mutation. |
|
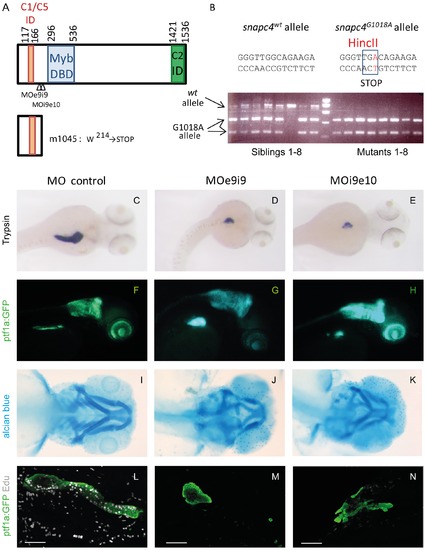
A nonsense mutation in the ORF of snapc4 is the m1045 causal mutation. A) Schematic scheme of the zebrafish Snapc4 protein. The conserved Myb DNA binding domain and the interacting domains for Snapc1/snapc5 and Snapc2 as described for the human SNAPC4 ([16], [17] are indicated by boxes. The m1045 mutation generates a premature stop codon in the zebrafish snapc4 cDNA resulting in a truncated protein of 213 aa. The positions targeted by the MOe9i9 and MOi9e10 snapc4 morpholinos are indicated with arrowheads. B) Genotyping of m1045 mutant and unaffected sibling embryos by RFLP analysis. C–N) Phenotype of embryos injected with 8 ng of control morpholino, 8 ng of MOe9i9 or 6 ng of MOi9e10 snapc4 morpholinos. C–E) Dorsal view of WISH performed with a trypsin probe of 3.5 dpf larvae: 84% of the control morphants display a complete extension of the pancreatic exocrine tail (n = 19) while 52% of MOe9i9 morphants show a drastic reduction of the pancreatic exocrine tail and 48% no extension at all (n = 19) and 9% of MOi9e10 morphants show a drastic reduction of the pancreatic exocrine tail and 91% no extension at all (n = 23). F–H) Lateral view of 3.5 dpf Ptf1:GFP larvae: 95% of the control morphants display a complete extension of the pancreatic exocrine tail (n = 25) while 13% of MOe9i9 morphants show a drastic reduction of the pancreatic exocrine tail and 87% no extension at all (n = 83) and 24% of MOi9e10 morphants show a drastic reduction of the pancreatic exocrine tail and 62% no extension at all (n = 55). I–K) Alcian blue staining of 4 dpf embryos: MOe9i9 and MOi9e10 morphants display a reduction and malformation of the hyoid concomitant with the absence of the branchial arches 3 to 7. L–N) Confocal projections of the pancreatic region of embryos injected with Edu at 95 hpf and fixed at 96 hpf. All the control morphants show a high Edu incorporation rate (n = 15). 95% of the MOe9i9 morphants did not incorporate any Edu at all (n = 19) and 43% of the MOi9e10 morphants display no incorporation at all and 21% a considerably reduced incorporation rate (n = 14). Scale bars : 50 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
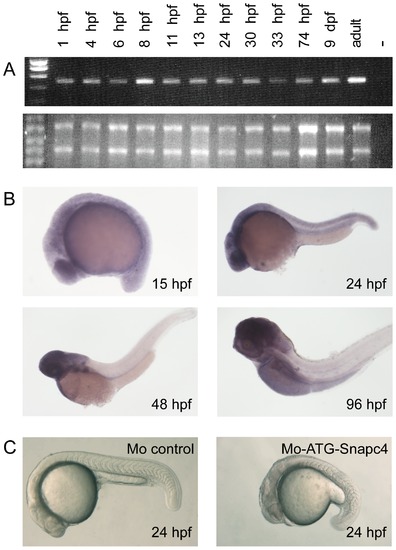
A: Time course analysis of zebrafish snapc4 expression by semi-quantitative RT-PCR. A control without addition of RNA (lane -) was included as a negative control. 3 μg of total RNA extracted at each time point were loaded on a denaturating agarose gel to check the quality and the quantity of the RNA used for each RT-PCR. B: Expression profile of the snapc4 transcripts performed by WISH at 15 hpf (12S), 24 hpf, 3 dpf and 4 dpf. C: Translation-blocking snapc4 morpholino leads to growth retardation before 24 hpf. Bright-field images of 24 hpf embryos injected with 8 ng of control morpholino or of MOATG-snapc4 : 75% of the embryos injected with MOATG-snapc4 show severe growth retardation compared to the control morphant. |
|
Genotyping of m1045 mutant and unaffected sibling embryos by RFLP analysis. |
|
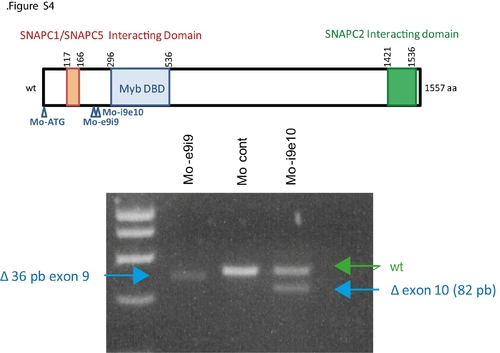
Control of the morpholino efficiency of the splicing-blocking morpholinos. RT-PCR analysis of total RNA extracted from 30 hpf morphants show that the snapc4 mRNA is truncated in the Moe9i9 and Moi9e10 morphants. |

Unillustrated author statements PHENOTYPE:
|