- Title
-
tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration
- Authors
- Kikuchi, K., Gupta, V., Wang, J., Holdway, J.E., Wills, A.A., Fang, Y., and Poss, K.D.
- Source
- Full text @ Development
|
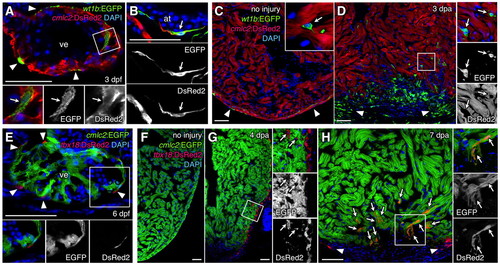
Zebrafish wt1b and tbx18 regulatory sequences drive expression in epicardial and non-epicardial tissue including cardiomyocytes. (A,B) wt1b:EGFP (green) is detectable in epicardial cells (arrowheads), as well as in ventricular (A) and atrial (B) cardiac muscle (arrows), where it colocalizes with cmlc2:DsRed2 (red) at 3 days post-fertilization (dpf). (C,D) wt1b-driven EGFP marks a subset of epicardial cells (arrowheads) in the uninjured adult ventricle, as well as intra-myocardial cells (inset, arrow) that do not express cmlc2:DsRed2 (C). At 3 days post-amputation (dpa), many wt1b:EGFP+ cells lacking cmlc2:DsRed2 are evident in and around the injury site (D). A subset of these cells are epicardial (arrowheads), whereas many are intra-myocardial (arrows). Higher magnification of the boxed area is shown in each channel on the right. An antibody was used to detect EGFP in A-D. (E) tbx18:DsRed2 (red) labels epicardial cells (arrowheads) at 6 dpf, but does not colocalize with cmlc2:EGFP (green). (F,G) tbx18:DsRed2 has little or no detectable expression in the uninjured adult ventricle (F), but is detectable after injury in epicardial cells and EPDCs (G, arrows), where it does not colocalize with cmlc2:EGFP. (H) At 7 dpa, tbx18:DsRed2 colocalizes with cmlc2:EGFP in many cardiomyocytes at the injury site (arrows). Arrowheads indicate epicardial tbx18:DsRed2 fluorescence. An antibody was used to detect DsRed in E-H. at, atrium; ve, ventricle. Scale bars: 50 μm. |
|
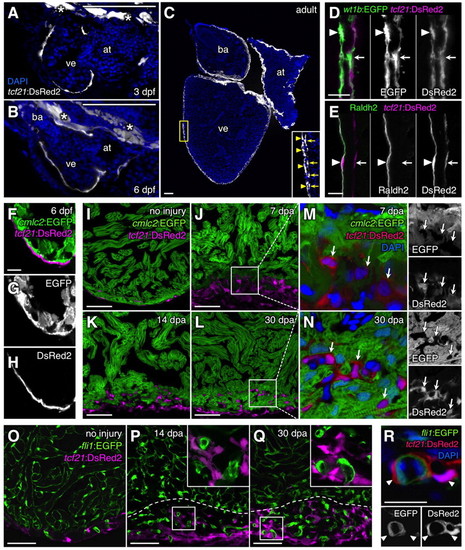
Zebrafish tcf21 regulatory elements drive epicardial-specific expression. (A-C) Cardiac tcf21:DsRed2 expression (white) at 3 dpf (A), 6 dpf (B) and adult (C) stages. Asterisks indicate extracardiac expression in A and B. Inset in C shows enlargement of area in yellow box. (D,E) tcf21:DsRed2+ epicardial cells (magenta) within the adult ventricular wall, assessed for wt1b:EGFP (D) or antibody staining for Raldh2 (E; green) colocalization. Arrowheads and arrows in C-E indicate the outer (epithelial) epicardial cell layer and EPDCs, respectively. (F-I) tcf21:DsRed2 (magenta) and cmlc2:EGFP (green) expression in distinct cells of 6 dpf (F-H) and adult (I) ventricles. An antibody was used to detect DsRed in F-H; this antibody also failed to detect DsRed2+EGFP+ cells in adult tcf21:DsRed2; cmlc2:EGFP ventricles (data not shown). (J-N) tcf21:DsRed2 (magenta or red) and cmlc2:EGFP (green) expression also mark distinct cells during regeneration. Arrows indicate tcf21:DsRed2+ nuclei in M and N. (O-R) tcf21:DsRed2 (magenta or red) and fli1:EGFP (green) do not colocalize in uninjured or regenerating adult ventricles. Insets in P and Q are enlarged views of boxed areas. Dashed lines indicate approximate amputation planes. at, atrium; ba, bulbus arteriosus; ve, ventricle. Scale bars: 100 μm for A-C; 10 μm for D-H,R; 50 μm for I-Q. |
|
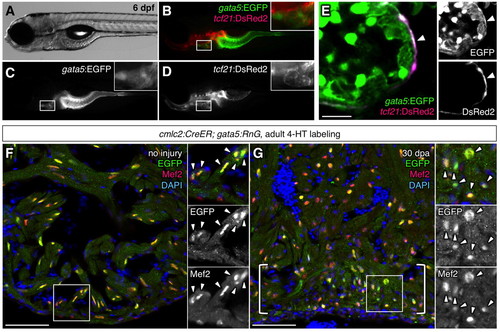
gata5 and tcf21 regulatory sequences drive expression that overlaps in the zebrafish epicardium. (A-E) A gata5:EGFP; tcf21:DsRed2 larva at 6 dpf. Insets are magnified images of the areas in white boxes in B-D. A single confocal slice visualizing gata5:EGFP; tcf21:DsRed2 ventricles shows colocalization limited to the epicardium (E, arrowhead). Each channel of the image is shown on the right (E). (F,G) The gata5:RnG indicator line efficiently reports induced Cre-mediated recombination in cardiomyocytes. cmlc2:CreER; gata5:RnG adults treated with 4-HT, showing EGFP labeling in Mef2+ cardiomyocytes nuclei of uninjured (F) and regenerated (G, brackets) cardiac tissue. Arrowheads in insets indicate EGFP immunofluorescence. Scale bars: 10 μm for E; 50 μm for F,G. |
|
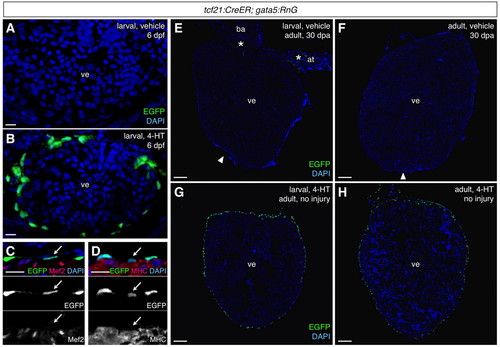
Efficient and specific induced epicardial labeling in tcf21:CreER; gata5:RnG zebrafish. (A,B) tcf21:CreER; gata5:RnG larvae incubated with vehicle (A) or 4-HT (B) at 3-5 days post-fertilization (dpf) and visualized at 6 dpf, indicating 4-HT-induced labeling of the larval epicardium (green). Confocal projections of 10 μm z-stacks are shown. (C,D) Confirmation of epicardial specificity at 6 dpf embryos, assessed by Myosin heavy chain (MHC) and Mef2 staining. Arrows indicate an epicardial cell nucleus in each image. (E,F) 30 days post-amputation (dpa) ventricles from adult tcf21:CreER; gata5:RnG animals treated with vehicle as larvae (E) or adults (F). Occasional epicardial labeling (E, asterisks) was observed in injured or uninjured hearts. Arrowheads indicate apical regenerate. (G,H) 4-HT treatment at larvae or adult stages specifically labeled the majority of adult epicardial cells. at, atrium; ba, bulbus arteriosus; ve, ventricle. An antibody was used to detect EGFP in these experiments. Scale bars: 10 μm for A-D; 100 μm for E-H. |
|
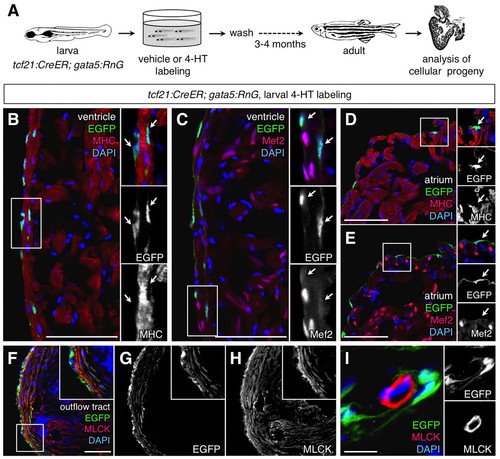
Adult progeny of labeled larval epicardial cells. (A) Schematic of the experimental design. (B,C) Portions of the ventricle from the lineage-traced heart, stained with an antibody for EGFP (green) and muscle markers (red). Lineage-labeled EGFP+ cells (arrows in inset) did not colocalize with cytosolic Myosin heavy chain (MHC; B) or nuclear Mef2 (C; n=10). The small EGFP+ region at the bottom of the inset of B is a thin strand of epicardial cytosol partially overlaying myocardial cytosol. A structure like this cannot be resolved by confocal imaging in processed tissue sections, but has neither the size nor morphology of a cardiomyocyte. Moreover, EGFP fluorescence was never observed in Mef2+ nuclei. (D,E) Portions of the atrium from the lineage-traced heart, stained with an antibody for EGFP (green) and muscle markers (red). Lineage-labeled EGFP+ cells (arrow in inset) did not colocalize with MHC (D) or Mef2 (E) (n=10). (F-I) EGFP+ cells in the outflow tract colocalize with Myosin light chain kinase (MLCK; red), a smooth muscle marker (F-H). This colocalization was not observed in coronary vascular smooth muscle (I). An antibody was used to detect EGFP in these experiments. Scale bars: 50 µm for B-H; 10 μm for I. |
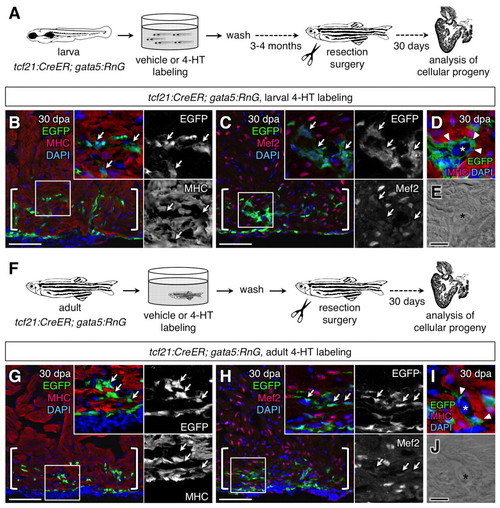
|
Epicardial cells contribute vascular support cells during heart regeneration. (A) Schematic of the experimental design. (B,C) Lineage-labeled EGFP+ cells (arrows, green) did not colocalize with Myosin heavy chain (MHC; B, red) or Mef2 (C, red) in 30 days post-amputation (dpa) regenerates of the larvally labeled animals (brackets; n=20). (D,E) EGFP+ cells (arrowheads) surrounds a vessel within the larvally labeled regenerate. Asterisk marks vascular lumen. (F) Schematic of the experimental design. (G,H) Lineage-labeled EGFP+ cells (arrows) did not colocalize with MHC (G) or Mef2 (H) in 30 dpa regenerates of the adult-labeled animals (brackets; n=6). (I,J) EGFP+ cells (arrowheads) surround a vessel within the adult-labeled regenerate. Asterisk marks vascular lumen. An antibody was used to detect EGFP in these experiments. Scale bars: 50 μm for B,C,G,H; 10 μm for D,E,I,J. |
|
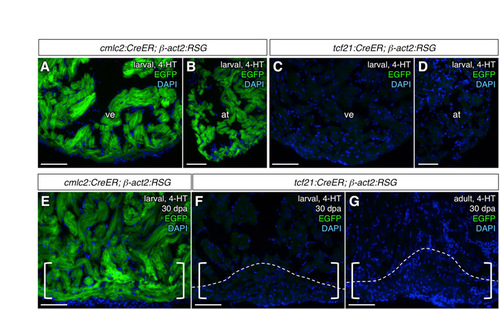
tcf21:CreER does not label cardiomyocytes or precursors of cardiomyocytes in the β-act2:RSG muscle indicator background. (A,B) Robust cardiomyocyte labeling (green) of adult ventricles and atria of cmlc2:CreER; β-act2:RSG animals treated as larvae with 4-hydroxytamoxifen (4-HT). (C,D) Ventricles and atria of adult tcf21:CreER; β-act2:RSG treated as larvae with 4-HT display no cardiomyocyte enhanced green fluorescent protein (EGFP) labeling. (E,F) 30 days post-amputation (dpa) regenerates (brackets) from cmlc2:CreER; β-act2:RSG (E) or tcf21:CreER; β-act2:RSG (F) animals that were treated with 4-HT at larval stages, assessed for cardiomyocyte EGFP labeling (n=6). (G) No cardiomyocyte EGFP labeling was observed in 30 dpa regenerates (brackets) of tcf21:CreER; β-act2:RSG that underwent 4-HT labeling as adults (n=4). Dashed lines indicate approximate amputation planes. at, atrium; ve, ventricle. Scale bars: 50 μm. |