- Title
-
CXCR4 and CXCR7 cooperate during tangential migration of facial motoneurons
- Authors
- Cubedo, N., Cerdan, E., Sapede, D., and Rossel, M.
- Source
- Full text @ Mol. Cell Neurosci.
|
Motoneurons express cxcr4b during tangential migration. (A, B) cxcr4b expression at 19 hpf (in situ hybridization). (A) Dorsal view: labeled cells are observed in r4, r5 and in r6 (dotted circle: otic vesicle). (B) Lateral view showing the ventral position of cxcr4b expressing cells. (C–E) Motoneurons co-express cxcr4b and Islet-1 during tangential migration, (double fluorescent in situ hybridization) on 24 hpf embryos. (C) cxcr4b is expressed by cells localized in rhombomere r3, r4–6. (D) Islet-1 expression in motoneurons and sensory ganglia (og: otic ganglion). (E) Merged image illustrating the co-localization of cxcr4b and Islet-1 expression (arrowhead: downregulation of cxcr4b expression). (F) cxcr4b expression (Nomarski image of in situ hybridization stainings which were inverted (red pseudocolor) and overlaid with GFP immunolabeling (G, green) shows cxcr4b co-expression in the migrating GFP positive cells. (H, merge image). Note in r4, cxcr4b cells with GFP expression (*). Scale bar: 50 μm. |
|
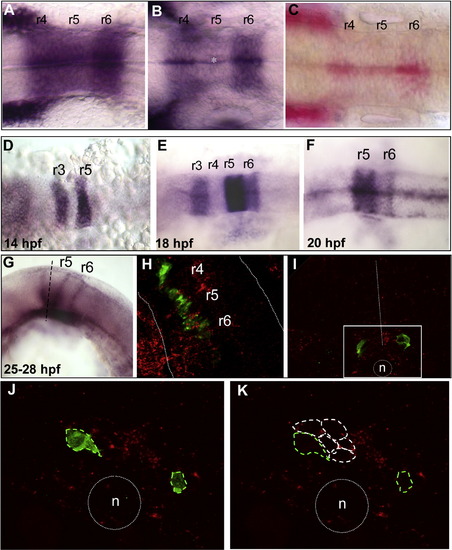
sdf1a and cxcr7b expression patterns during hindbrain development. sdf1a expression (in situ hybridization) (A–C). (A) At 14 hpf, sdf1a is strongly expressed in r4, r5 and r6. (B) At 18 hpf, expression has decreased in r5 (*) and becomes almost restricted to the midline at 20 hpf (C, Fast red substrate). cxcr7b expression (D–G). (D) At 14 hpf cxcr7b is expressed in r3 and r5. (E) At 18 hpf, a strong expression is maintained in r5, but expression decreases in r3 and appears in r6. (F) By 20 hpf, expression has disappeared from r3 and decreased in r6. (G) At 24 hpf the boundaries of r5 and r6 are labeled as well as the ventral part of r5. (H) Confocal single merge image at 24 hpf (double fluorescent in situ hybridization), the detection of cxcr7b (red) and of cxcr4b (green) reveals that cxcr4b expressing cells are localized through cxcr7b expressing area (dotted white line outlined hindbrain borders). (I) Transverse section through r5 after cxcr7b in situ hybridization (red) combined with GFP immunolabeling (green). Migrating cells (GFP-positive) are localized in cxcr7b positive region (white square area), dotted white line = midline, n = notochord. (J, K) High magnification of single confocal optic section shows exclusive expressions: GFP-positive cells (green dotted line) are devoided of cxcr7b expression, which is expressed in the juxtaposed cells (white dotted line). |
|
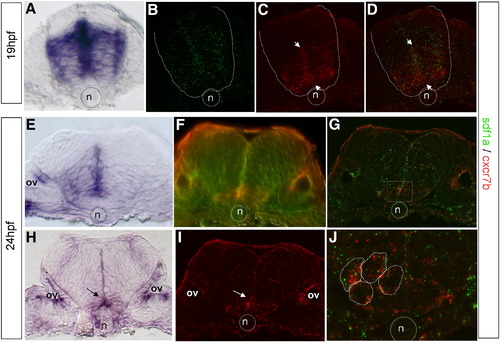
Co-expression of CXCR7 and SDF1 is restricted to a sub-population of neuroepithelial cell in r5. (A) Cross-sections at the level of r5 level at 19 hpf. Expression of sdf1a is intense in the germinal zone around the midline and is absent both in the dorsal region of the neural tube and in the floorplate. (B–D, F, G, I, J) Double insitu hybridization of sdf1a (green, B, F, G, J) and cxcr7b (red, C, F, G, I), the dotted line delimited the neural tube border and the dotted circle the notochord position. (C) cxcr7b is prominent in the ventral area of r5. The merge image indicates a co-expression in the ventral half of r5, arrows (D). (E–J) Expression patterns at 24 hpf. (E) Cross-section at the r5/r6 boundary (24 hpf) reveals sdf1a expression in the basal plate, most prominently within the periventricular region, with exclusion from the floor plate, arrows. (F) Double FISH after observation with Nomarski optic, which allow cell border visualization or confocal microscopy analysis (G). (I, J) Detection of cxcr7b (red) and sdf1a (green, arrows) in r5. (H, I) Cross-section through r5 shows that cxcr7b expression is restricted to a few cells adjacent to the floor plate, arrow. (J) Single merge image of a confocal Z-stack showing that both genes are expressed within the same cells (underlined, red and green spots). n = notochord, circle, ov = otic vesicle. |
|
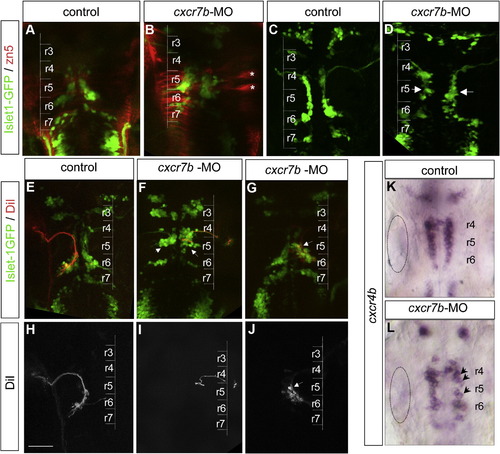
cxcr7b inactivation leads to a defect of facial motoneuron migration. (A, B) zn5 immunolabeling assess GFP cell position in control (A) and cxcr7b-MO (B), zn5 is considered as a good marker for commissural neurons. r5 commissural neurons are particularly prominent (asterisks). (C, D) At 24 hpf, Islet-1-GFP expressing motoneurons form a smooth stream in control embryos but are less well organized and begin to accumulate in r5 in cxcr7b morphant (D) embryos (arrows). (E, J) Retrograde DiI labeling of facial motoneurons (red) in an Isl1-GFP background (green) at 48 hpf. The lower panels show the DiI labeling alone (H–J). (E, H) In control embryos DiI labeled cells are located mostly in r6. (F, I) In cxcr7b morphant embryos where GFP cells are positioned mainly in r5 (severe phenotype, arrows in F), DiI labeled cells are mostly found in r5. (G, J) In morphant embryos with a milder phenotype, GFP cells distributed within r5–r6 (G, arrow), DiI labeled cells are predominantly found in r5 (J). Inactivation of cxcr7b does not affect cxcr4b expression level. (K, L) The level of cxcr4b expression is similar in normal (K) and morphant (L) embryos but the pattern is disorganized in the morphants (arrows dotted circle: otic vesicle. Scale bar: 50 μm. |
|
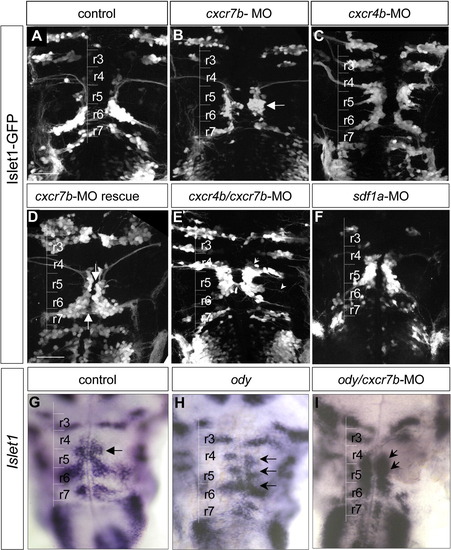
CXCR7b and CXCR4b are required for proper motoneuron migration. (A) In a control 48 hpf embryo, Isl1-GFP positive cells accumulate in r6. (B) ln cxcr7b-MO, most Isl1-GFP positive cells accumulate in r5 (arrow). (C) In cxcr4b-MO embryo, Isl1-GFP positive cells are present in r4 and r5, reflecting a partial impairment of migration. (D) In cxcr7b-mRNA injected (600 pg) embryos, a partial rescue is observed with no more accumulation in r5 but many Isl1-GFP cells remained close to the midline from r4 to r6 (arrows). (E) Double inactivation of cxcr4b and cxcr7b by MO results in facial migration defects compared to control Islet-1-GFP embryos (A). The most severe phenotype was characterized by GFP positive cell accumulation in r4 and r5 and an almost complete absence of cells in r6. Ectopic dendrites can be seen contralaterally (arrow) or toward the periphery (arrowhead). (F) A similar phenotype is observed in sdf1a-MO embryos: few cells are visible in r6 while motoneurons accumulate in r4 and r5. (G–I) Double ody/cxcr7b-MO embryos at 42 hpf (I) were compared to wild type (G) and to ody embryo (H) using Islet-1 in situ hybridization. In control, Islet-1 is expressed by facial motoneurons in r6 and by the abducens nucleus in r5 (arrow, G). Note that cells accumulate mainly in r4, r5 and r6 in ody mutant (H, arrows) while in ody/cxcr7b-MO Islet-1 cells are found in r4 and r5 with few cells in r6 (I, arrows), similar to the double morphant embryos (E). In all comparisons, control and mutant embryos were age-matched. Scale bar: 50 μm. |
|
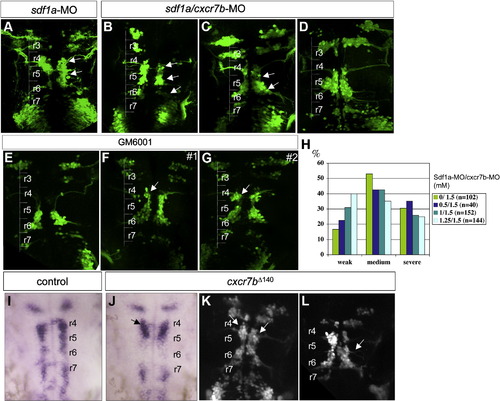
Double sdf1a/cxcr7b inactivation partially restored motoneuron migration (A,D). Compared to sdf1a phenotype (A), three examples of double morphants are given: a severe case (B) which is comparable to (A) with cells arrested in r4 and r5 (arrows) and a weak case (C), which corresponds to the major phenotype: cells are retrieved in r4, r5, r6 (B, C embryos were injected at the ratio sdf1a-MO/cxcr7b-MO 1.25/1.5). (D) Typical mixed phenotype with r5 accumulation on the left side (CXCR7b phenotype) and cell accumulation along the midline on the right side with ectopic dendrites (injection at the ratio sdf1a-MO/cxcr7b-MO 0.5/1.5). H: graphic illustration of the phenotype variability from a range of concentration of sdf1a-MO (0-1.25 mM) with constant cxcr7b-MO concentration (1.5 mM). Three categories have been distinguished, the severe phenotype includes strong phenotypes of cxcr7b-MO and sdf1a-MO. The Y axis corresponds to the percentage of embryos in each group. (E–G) Motoneuron migration at 48 hpf after GM6001, MMP inhibitor, ventricular injections of Isl1-GFPembryos (24 hpf). GM6001 injected embryos (n = 25) showed a variability in cell arrest position in r4 (F, G, arrows). (G) #2 embryo displays the more severe phenotype observed (n = 5/25). No specific arrest in r5 ever occurs. (I–K) Ectopic expression of cxcr7b truncated receptor cxcr7bΔ140 by mRNA injection. cxcr4b expression is not altered in mRNA cxcr7bΔ140 injected embryos (J), however it reflects a failure of the migration of CXCR4b expressing cells with accumulation in r4 (arrow) at 24 hpf. (K) At 48 hpf, injected Islet1-GFPembryos display a migration defect with cell accumulation in r4 and r5 (K, arrows) and in most severe cases, ectopic dendrites (L, arrow). cxcr7bΔ140 mRNA injection was performed at 40 pg (J, K) and 80 pg (L). |
|
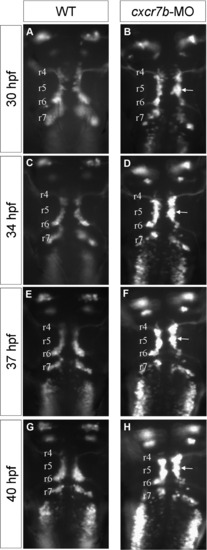
Time-lapse analyses of facial motoneuron phenotype in cxcr7b-MO embryo (dorsal view). Four time points from time-lapse observations during facial motoneuron migration on Islet-1-GFP wild type (A,C,E,G) and cxcr7b morphant (B,D,F,H) embryos. The time-lapse movie extends from 28 to 42 hpf and the time-points were 30 pf (A,B), 34pf (C,D), 37 pf (E,F) and 40 pf (G,H).The total length of the hindbrain shortens during this period of time, and the correspondence between the otic vesicle and r5 is progressively lost, however the position where a large number of cells are arrested in the morphant is easily followed during the time-lapse (arrow) and can be traced back to r5. |
|
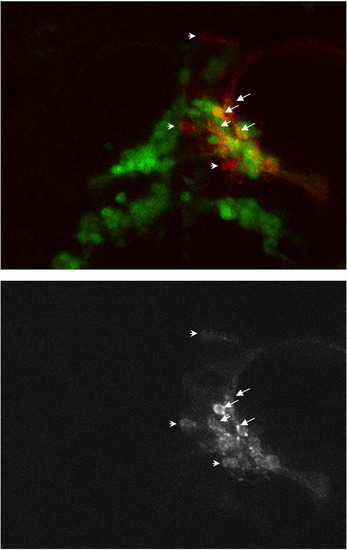
High magnification of Fig. 5C. The DiI labeling (red coincides with GFP positive cells in most cases (arrows) although 3 cells are clearly GFP negative (arrowheads). |
|
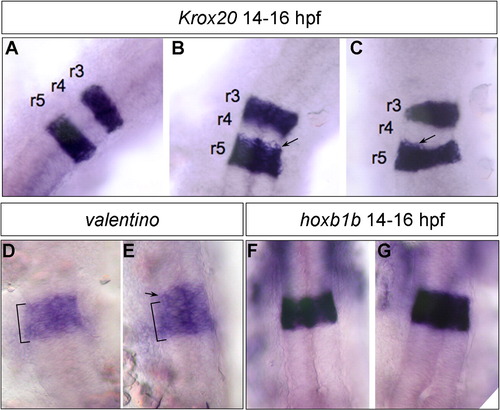
Normal rhombomere specification in cxcr7b-MO. Segmentation pattern of the hindbrain was examined with Krox20 (A–C), valentino (D–E) and hoxb1a (F–G). In most cases expression of the segmentation genes was normal, except in 30% of the injected embryos where the rhombomeric limits were irregular with a mild extension in Krox20 (B–C, arrows) and val (E, arrow) domains (n = 150). hoxb1a expression was not detectably affected (compare F and G) (n = 230). |
|
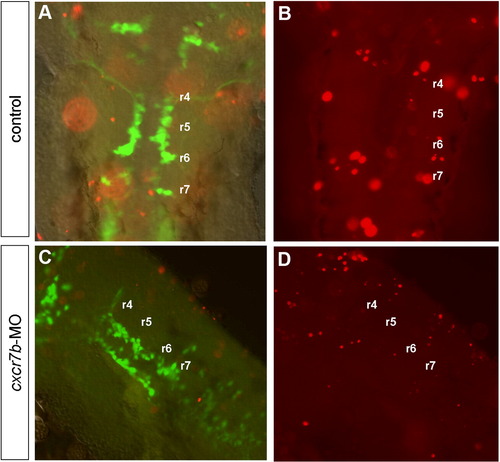
Cell death analysis using the TUNEL method (red) in control (A,B) and in cxcr7b-MO (C,D) embryos at 28–30 hpf. (A,B) A few apoptotic cells are observed in control embryos in the dorsal part of the hindbrain, scattered from r2 to r7. (A,C) Merge images of GFP and TUNEL labeling at the level of facial motoneuron migration show that GFP cells have a ventral position while apoptotic cells have a dorsal position in control (A) as well as cxcr7b-MO embryo (C). (B,D) Apoptotic cells are in focus at the level of dorsal hindbrain. (C,D) No labeling was observed predominantly localized in r3 or r5 in cxcr7b-MO. |
Reprinted from Molecular and cellular neurosciences, 40(4), Cubedo, N., Cerdan, E., Sapede, D., and Rossel, M., CXCR4 and CXCR7 cooperate during tangential migration of facial motoneurons, 474-484, Copyright (2009) with permission from Elsevier. Full text @ Mol. Cell Neurosci.