- Title
-
Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis
- Authors
- Kosaka, K., Kawakami, K., Sakamoto, H., and Inoue, K.
- Source
- Full text @ Mech. Dev.
|
The distributions of germ plasm RNAs during oogenesis. In situ hybridization of serial sections of the zebrafish ovary. Distributions of vasa and dazl RNAs (a, b, e, f, i, j, m and n) and nanos1 and dazl RNAs (c, d, g, h, k, l, o and p) are compared in the serial sections of oocyte at stages I (a–h), II (i–j), and III (m–p). Sections were cut at a thickness of 10 μm. Scale bars, 50 μm EXPRESSION / LABELING:
|
|
Co-localization of germ plasm RNAs with the mitochondrial cloud in zebrafish oocytes. (A) Stage I oocyes were stained with Mitotracker Red580 (mitochondria) and DAPI (DNA). Schematic representation was shown at the bottom of each panel. In panels a and b, a mitochondria cloud was found adjacent to the germinal vesicle (GV), whereas mitochondria staining was observed at or near the vegetal cortex in panel c. (B) The right panel shows mitochondria staining of a section of a stage I oocyte. For the left panel, in situ hybridization was carried out on the same section as that shown in the right panel using the vasa, nanos1, zDazl and zorba/ZOR-1 probes. Sections were cut at a thickness of 10 μm. Scale bars, 50 μm. EXPRESSION / LABELING:
|
|
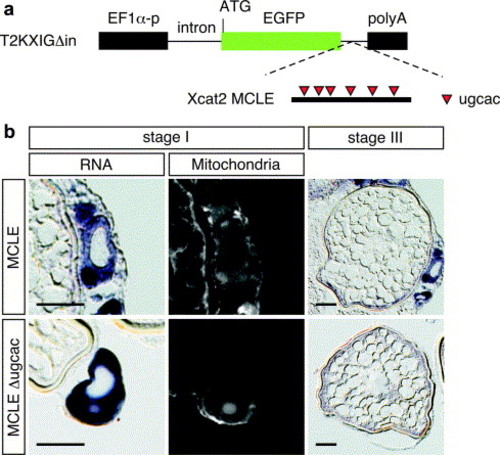
The Xcat2 3′ UTR directs localization to the mitochondria cloud in transgenic zebrafish oocytes. (a) Schematic representation of the transgenic construct used to express a GFP mRNA fused with Xcat2 3′ UTR. Xcat2 MCLE or Xcat2 MCLEΔugcac 3′ UTR (Betley et al., 2002) was inserted into the downstream site of EGFP in the T2KXIGΔin plasmid vector (Kawakami et al., 2004). Expression of GFP mRNA is driven by the EF1α promoter. (b) In situ hybridization in transgenic zebrafish oocytes expressing GFP mRNA fused with Xcat2 MCLE (upper panel) or MCLEΔugcac (lower panel), using antisense GFP RNA as a probe. Left column, expression of the reporter mRNA in a stage I oocyte; middle column, the same oocyte counterstained with Mitotracker. Right column, expression of the reporter mRNA in a stage III oocyte. Sections were cut at a thickness of 10 μm. Scale bars, 50 μm. |
|
Hermes protein is a constituent of the mitochondrial cloud. (a) Schematic representation of Hermes proteins in zebrafish and Xenopus (Gerber et al., 1999). RRM represents an RNA-recognition motif. The positions of conserved motifs in RRM, RNP2 and RNP1 (Birney et al., 1993) are indicated. Amino acid sequence identity among Hermes proteins in the N-terminal RRM and C-terminal regions is shown. Numbering according to amino acids in the Hermes proteins. (b) Immunodetection of Myc-tagged zebrafish Hermes using an antibody against Xenopus Hermes. The in vitro synthesized mRNA encoding Myc-zHermes was injected into 1-cell embryos and whole lysates of injected embryos were collected at 12 hpf (I). Control lysates from uninjected embryos are shown in (U). Two embryo equivalents per lane were loaded onto the gel. Immunoblotting was done with anti-Hermes and anti-Myc antibodies. As expected, the Myc-Hermes protein was detected at approximately 30 kDa. (c) Immunoblot analysis of endogenous Hermes protein in early stage zebrafish embryos (at 1-cell and 4-cell stages). (d) Distribution of dazl RNA (left) and zHermes protein (right) in the serial sections from stage I and II oocytes (upper and lower panels, respectively). Sections were cut at a thickness of 10 μm. Scale bars, 50 μm. |
|
The 3′ UTRs of vasa, nanos1 and dazl RNAs govern localization during oogenesis and early embryogenesis. In situ hybridization of GFP mRNA fused with vasa, nanos1, dazl, or SV40 3′ UTRs expressed in transgenic zebrafish oocytes (stages I and III) and fluorescein in situ hybridization in 4-cell embryos. GFP-nanos1 mRNA was expressed by zpc promoter (Onichtchouk et al., 2003), whereas other mRNAs were expressed by EF1α promoter. In stage I oocytes, in situ hybridization (RNA) was performed on sections counter-stained with Mitotracker (Mitochondria). Oocyte sections were cut at a thickness of 10 μm. Scale bars, 50 μm. Lateral and animal views of the 4-cell embryo are shown. |
|
Identification of cis-elements required for localization of dazl mRNA. (a) Schematic representation of deletions of zebrafish dazl 3′ UTR (Δ1–Δ9) used in transgenic experiments. Numbers represent nucleotide positions relative to the stop codon. Regions of the dazl 3′ UTR labeled a, b, c and d contain nucleotides 1-199, 200-375, 376-782, and 783-1218, respectively. The results of mRNA distribution analyses are indicated to the right of the schematic diagram. (+), normal pattern of distribution; (-), not localized. MC shows MC localization in oocytes. Vg shows localization at vegetal pole in 4-cell embryos. (b) Detection of the GFP mRNA fused with mutant 3′ UTRs in transgenic oocytes (stages I and III) and 4-cell embryos. For stage I oocytes, in situ hybridization was performed on sections counterstained with Mitotracker to detect mitochondria. Oocyte sections were cut at a thickness of 10 μm. Scale bars, 50 μm. A lateral view of the 4-cell embryo is shown. |
|
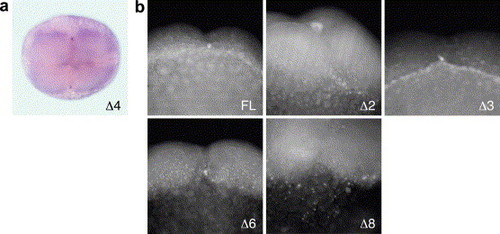
Distribution of a reporter mRNA fused with mutant dazl 3′ UTRs in 4-cell embryos. (a) In situ hybridization of Δ4 RNA. Animal view of the 4-cell embryo shown in Fig. 6b. (b) Fluorescein in situ hybridization in transgenic 4-cell embryos expressing GFP mRNA fused with full-length (FL), Δ2, Δ3, Δ6, or Δ8 dazl 3′ UTRs. The furrow region is enlarged. |

Unillustrated author statements |
Reprinted from Mechanisms of Development, 124(4), Kosaka, K., Kawakami, K., Sakamoto, H., and Inoue, K., Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis, 279-289, Copyright (2007) with permission from Elsevier. Full text @ Mech. Dev.