- Title
-
Mimicry Embedding Facilitates Advanced Neural Network Training for Image-Based Pathogen Detection
- Authors
- Yakimovich, A., Huttunen, M., Samolej, J., Clough, B., Yoshida, N., Mostowy, S., Frickel, E.M., Mercer, J.
- Source
- Full text @ mSphere
|
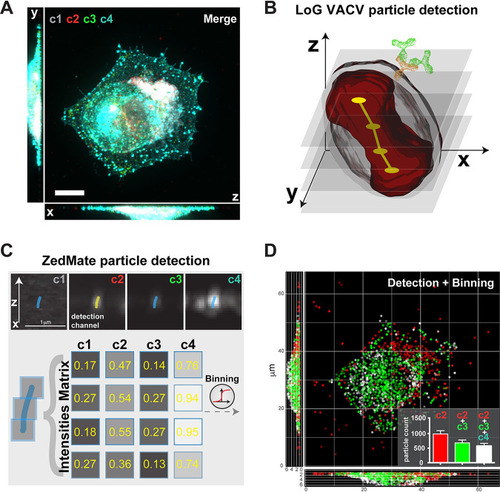
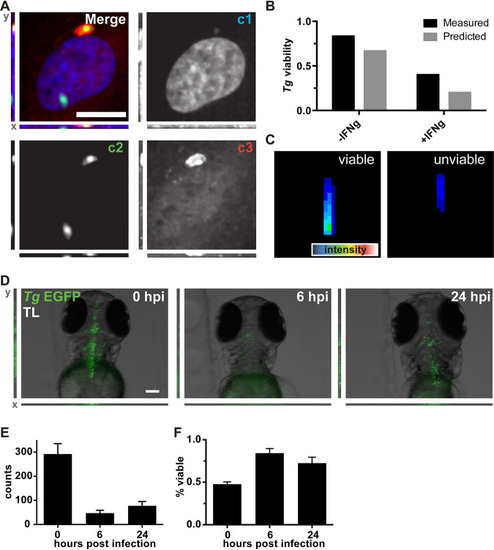
ZedMate facilitates detection and classification of VACV particles in infected cells. (A) Merged four-channel fluorescent image of a HeLa cell infected with VACV (see |
|
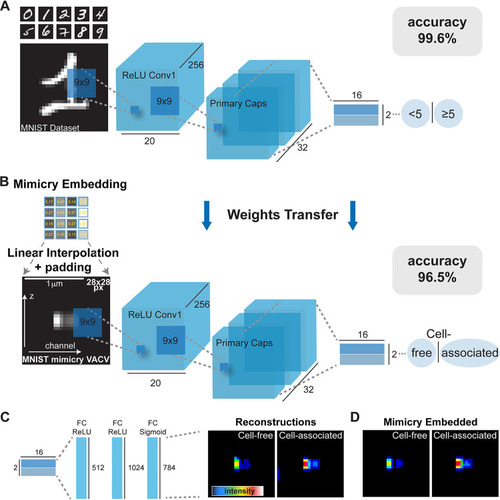
Mimicry embedding allows separation of cell-free and cell-associated VACV particles through weights transfer from a CapsNet trained on the binary MNIST data set. (A) CapsNet architecture for training on the MNIST handwritten digits data set repurposed into a binary classification problem (<5 or ≥5) prior to CapsNet weights transfer. Black numbers represent dimensions of tensors. ReLU, rectified linear unit. (B) Mimicry embedding of VACV Z-profiles detected by ZedMate. The intensity matrix of fluorescence signal ( |
|
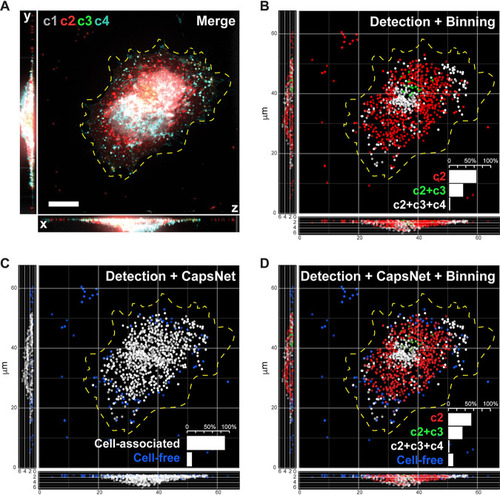
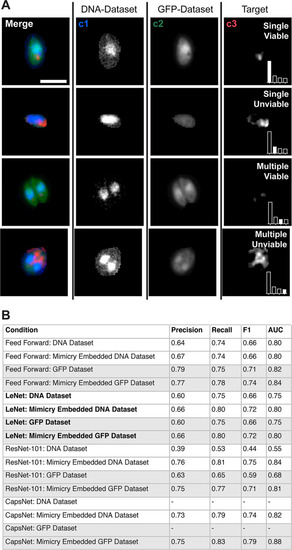
Inference demonstrates that mimicry embedding and trained CapsNet allow efficient classification of VACV particles into four biological classes. (A) Merged four-channel fluorescent image of a HeLa cell infected with VACV previously unseen by CapsNet (see |
|
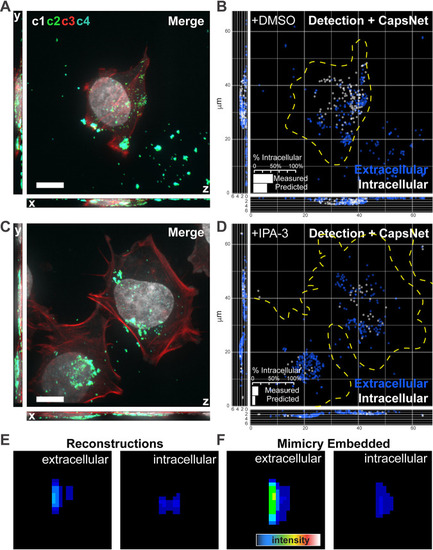
Mimicry embedding can be used for weak-labeling particle classification. (A) Merged four channel fluorescent image of a HeLa cell infected with VACV previously unseen by CapsNet (see |
|
Mimicry embedding and weight transfer employed for |
|
2D |