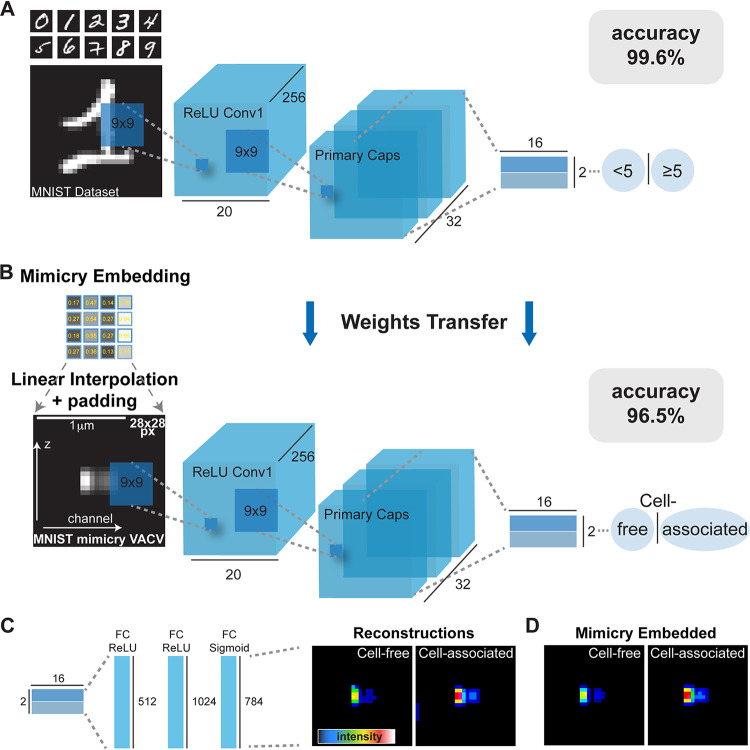
FIG 2
Mimicry embedding allows separation of cell-free and cell-associated VACV particles through weights transfer from a CapsNet trained on the binary MNIST data set. (A) CapsNet architecture for training on the MNIST handwritten digits data set repurposed into a binary classification problem (<5 or ≥5) prior to CapsNet weights transfer. Black numbers represent dimensions of tensors. ReLU, rectified linear unit. (B) Mimicry embedding of VACV Z-profiles detected by ZedMate. The intensity matrix of fluorescence signal (