- Title
-
The maternal to zygotic transition regulates genome-wide heterochromatin establishment in the zebrafish embryo
- Authors
- Laue, K., Rajshekar, S., Courtney, A.J., Lewis, Z.A., Goll, M.G.
- Source
- Full text @ Nat. Commun.
|
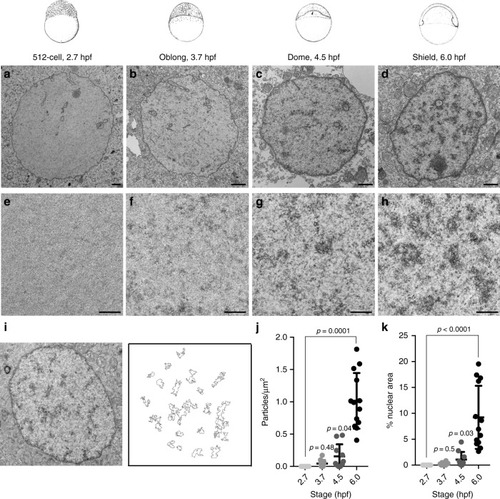
Chromatin of the early zebrafish embryo lacks H3K9me3. |
|
The early zebrafish embryo lacks condensed chromatin ultrastructure. Zebrafish embryonic stage schematics are reproduced from Kimmel et al. |
|
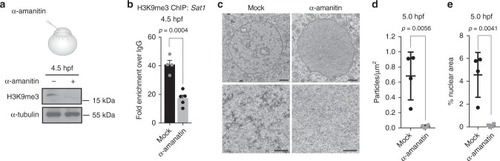
Blocking zygotic transcription impairs heterochromatin. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|
|
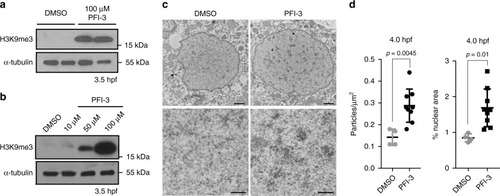
Smarca2 inhibition accelerates the timeline of chromatin compaction. |
|
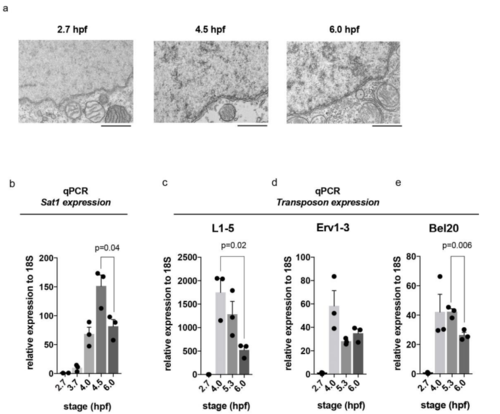
Elevated repetitive transcripts at stages with less condensed chromatin ultrastructure (a) TEM images showing nuclear periphery and cytoplasmic organelles at 2.7, 4.5 and 6.0 hpf. Scale bar equals 1 m. (b-e) Quantitative RT-PCR showing increasing levels of transcripts derived from repetitive elements following the onset of zygotic transcription. A decline in transcript levels at 6.0 hpf correlates with the onset of heterochromatin formation. All p-values were calculated using the students t-test, error bars indicate SEM. |
|
Abnormal development following Smarca2 depletion or inhibition (a) Bright field images of 24 hpf zebrafish larvae following injection with 2 ng of morpholinos targeting Smarca2 (MO1 or MO2) or 100 M PFI-3 at the 1 cell stage. Scale bars represent 1 mm. (b) Bright field images of 48 hpf zebrafish larvae following injection with 2 ng of morpholinos targeting Smarca2 (MO1 or MO2) or 100 M PFI-3 at the 1 cell stage. Scale bars represent 1 mm. (c) Quantification of embryos that were normal, dead or showed anomalies consistent with panel a, at 24 hpf. A minimum of 70 embryos were scored per condition. Numbers are representative of three independent experiments. |