Fig. 4
- ID
- ZDB-FIG-250728-111
- Publication
- Zhang et al., 2025 - IGF2BP1 restricts the induction of human primordial germ cell fate in an m6A-dependent manner
- Other Figures
- All Figure Page
- Back to All Figure Page
|
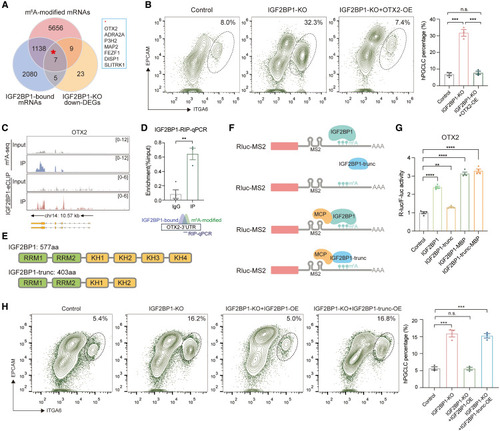
OTX2 mRNA is regulated by m6A modifications to restrict germ cell fate (A) Venn diagram showing the overlap of m6A-modified mRNAs, IGF2BP1-bound mRNAs, and downregulated DEGs (|log2FoldChange| > 2) in IGF2BP1-KO hESCs. (B) Flow cytometry showing the percentage of day-4 hPGCLCs induced from control, IGF2BP1-KO, and IGF2BP1-KO+OTX2-OE hESCs (mean ± SEM, n = 3, biological replicates; n.s., not significant, ∗∗∗p < 0.001, unpaired t test). (C) Tracks showing the m6A-seq peaks and IGF2BP1-eCLIP peaks on OTX2 genomic region. (D) RIP with antibodies against FLAG using IGF2BP1-3xFLAG knockin hESCs. qPCR was performed to determine the binding of IGF2BP1 to the 3′UTR of OTX2. Immunoglobulin G (IgG) served as a control. Enrichment of the indicated genes was normalized to the input level (mean ± SEM, n = 3, technical replicates; ∗∗p < 0.01, unpaired t test). (E) Schematic illustration of full-length and truncation (trunc) forms of IGF2BP1 protein. (F) Schematic illustration of tethering experiments for IGF2BP1 and IGF2BP1-trunc to the 3′UTR of OTX2. (G) Luciferase assay for detecting the activity of IGF2BP1, IGF2BP1-trunc, IGF2BP1-MCP, and IGF2BP1-trunc-MCP on OTX2-3UTR (mean ± SEM, n = 4, technical replicates; ∗∗p < 0.01, ∗∗∗∗p < 0.0001, unpaired t test). (H) Flow cytometry showing the percentage of day-4 hPGCLCs induced from control, IGF2BP1-KO, IGF2BP1-KO+IGF2BP1-OE, and IGF2BP1-KO+IGF2BP1-trunc-OE hESCs (mean ± SEM, n = 3, biological replicates; n.s., not significant, ∗∗∗p < 0.001, unpaired t test). See also Figure S4 and Table S3 |