Fig. 1
- ID
- ZDB-FIG-250528-19
- Publication
- Kaus-Drobek et al., 2025 - From discovery to potential application: engineering a novel M23 peptidase to combat Listeria monocytogenes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
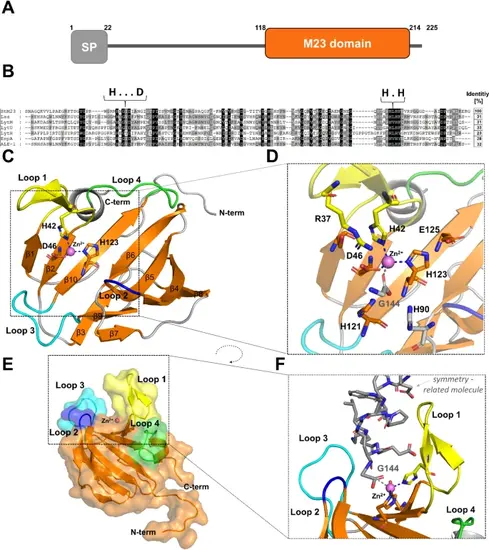
Bioinformatic and structural analysis of StM23 protein (PDB ID:9GY1). (A) Domain composition of the native M23/37 peptidase identified in S. thermophilus NCTC 10353, highlighting key structural features.(B) Sequence conservation analysis of M23 peptidases, showing the sequence identity of StM23 compared to other related peptidases. Zinc-binding motifs HxxxD and HxH are marked. (C) The overall fold of the M23 peptidase with characteristic structure of two beta sheets (orange) and distinctive loops: loop 1 (yellow), loop 2 (blue), loop 3 (cyan), and loop 4 (green), with residues involved in zinc binding highlighted. Zinc ion is shown as a purple sphere. (D) A closer view of the active site architecture, emphasizing residues that play a crucial role in catalytic function and active site stabilization. (E) Surface representation of the StM23 peptidase structure, illustrating the characteristic groove and the zinc ion (Zn²⁺) position in the groove. (F) Close-up view of the groove with zinc ion coordination, highlighting the role of Gly144 (depicted in grey) in zinc coordination. |