Fig. s4
|
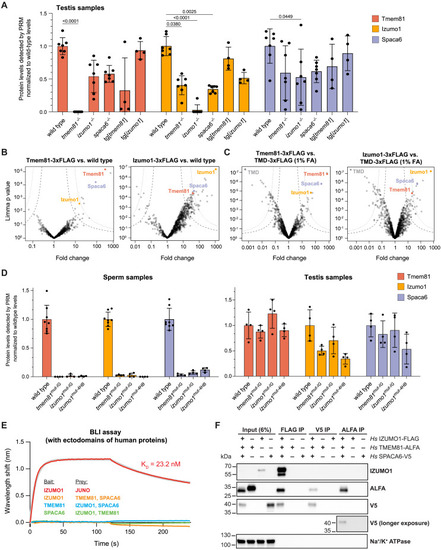
Experimental evidence for the interaction of Izumo1, Spaca6, and Tmem81 in zebrafish sperm and upon expression of the full-length human proteins in vitro, related to Figure 3 (A) Parallel reaction monitoring (PRM)42 mass spectrometry quantification of detected protein levels of Tmem81 (red), Izumo1 (yellow), and Spaca6 (blue) in wild-type and mutant zebrafish testis. Data were normalized by the loaded protein amount determined by 8 constant proteins and are shown relative to the mean wild-type level of each trimer member. In the case of Spaca6, 4 out of 5 peptides used for scheduled PRM (sPRM)-based quantification of Spaca6 protein levels lie upstream of the frameshift introduced in our spaca6−/− mutant, which is in line with the observed detection of these 4 peptides in spaca6−/− testis samples (but not sperm samples, where the C-terminally truncated Spaca6 is degraded). Of note, the 5th peptide, which lies downstream of the frameshift introduced in our spaca6−/− mutant, was neither detected in testis nor sperm samples, confirming the spaca6−/− genotype. Significant p values calculated by Kruskal-Wallis test with Dunn's mutiple comparisons test. All other comparisons are non-significant. (B and C) Volcano plots of differentially enriched proteins in FLAG coIPs from Tmem81-3xFLAG-sfGFP- or Izumo1-3xFLAG-sfGFP-expressing sperm versus wild-type sperm (B) or transmembrane domain (TMD)-3xFLAG-sfGFP-expressing sperm (C). Sperm samples in (B) were not crosslinked; sperm samples in (C) were lysed after crosslinking with 1% formaldehyde (FA) similar as shown in Figures 3B and 3C. (D) PRM mass spectrometry quantification of detected protein levels of Tmem81 (red), Izumo1 (yellow), and Spaca6 (blue) in wild-type and interface mutant zebrafish sperm (left) or testis (right). Those measured peptides that contained mutated residues were excluded from the analysis. (E) Biolayer interferometry (BLI) measurements to detect interactions between purified recombinant ectodomains of human trimer members. Four reciprocal BLI sensorgrams were measured: (1) positive control: biotinylated IZUMO1 as bait with association of 0.5 μM JUNO analyte (red curve), fitted curve (gray curve) was superimposed, (2) biotinylated IZUMO1 as bait with association of 5 μM TMEM81 and SPACA6 (orange curve), (3) biotinylated TMEM81 as bait with association of 5 μM IZUMO1 and SPACA6 (blue curve), and (4) biotinylated SPACA6 as bait with association of 5 μM IZUMO1 and TMEM81 (green curve). A single representative sensorgram was selected from triplicate measurements and shown here. (F) Western blot of human full-length trimer coIPs, including the reciprocal ALFA and V5 IPs from solubilized GPMVs from cells co-expressing Hs IZUMO1-3xFLAG, Hs SPACA6-3xV5, and Hs TMEM81-ALFA or expressing the individual proteins. Note that this western blot shows the complete results of the experiment described in Figure 3D, where only the input and FLAG IP are shown. The plasma membrane protein Na+/K+ ATPase is controlling for non-specific capture of membrane fragments. One western blot from three independent replicates is shown. Bar graphs represent mean ± SD. |
Reprinted from Cell, 187(25), Deneke, V.E., Blaha, A., Lu, Y., Suwita, J.P., Draper, J.M., Phan, C.S., Panser, K., Schleiffer, A., Jacob, L., Humer, T., Stejskal, K., Krssakova, G., Roitinger, E., Handler, D., Kamoshita, M., Vance, T.D.R., Wang, X., Surm, J.M., Moran, Y., Lee, J.E., Ikawa, M., Pauli, A., A conserved fertilization complex bridges sperm and egg in vertebrates, 7066-7078.e22, Copyright (2024) with permission from Elsevier. Full text @ Cell