Fig. 3
- ID
- ZDB-FIG-240807-3
- Publication
- Fuentes et al., 2024 - Maternal regulation of the vertebrate oocyte-to-embryo transition
- Other Figures
- All Figure Page
- Back to All Figure Page
|
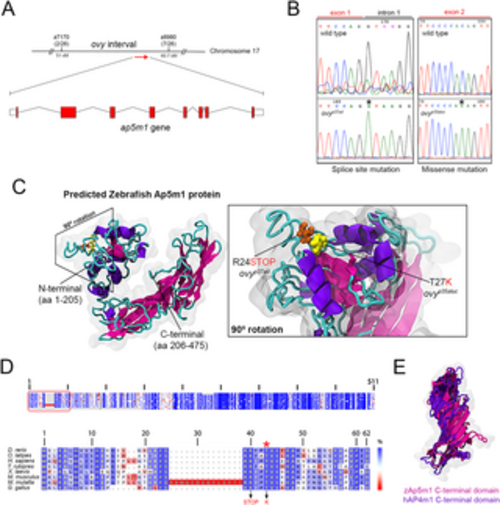
Molecular nature of the ovy gene. A. Genetic and physical map of the ovy locus and schematic representation of the zebrafish ap5m1 gene, which consists of 8 coding exons and 7 introns. z7170 and z8980 are SSLP markers flanking the ovy mutation. In parenthesis, the number of recombinants/total analyzed meioses defining the interval of ovy. Exons are shown as red boxes and introns as black lines. Sizes are not to scale. B. DNA sequencing analysis of the ovyp37ad and ovyp35aluc mutant alleles. Left: genomic DNA sequence of the wild-type and ovyp37ad allele indicates a single point mutation in the splice donor site of intron 1 of the ap5m1 gene (red rectangle). Right: cDNA sequence of the wild-type and p35aluc allele indicates a single point mutation in exon 2 of the ap5m1 gene (red rectangle). C. Left: predicted tertiary structure of the zebrafish Ap5m1 protein. The α- and β-helixes are colored in purple and pink, respectively, and the connecting loops in cyan. N- and C-terminal domains are indicated. The boundary of the two domains is at residue 205 (arrow head), which is found in the flexible loop comprised by residues 204–208. The approximate volumetric density map of the protein is shown in transparent gray. Right: predicted domain architecture of the Ap5m1 N-terminal portion containing residues Arginine 24 (R24) and Threonine 27 (T27). The amino acid change and premature stop codon caused by the ovyp35aluc and ovyp37ad mutations, respectively, are shown. D. Multiple amino acid alignments of the Ap5m1 protein and representative members of the vertebrate lineage. Top: Schematic of the protein alignment. The overall percentage (%) identity decreases from top to bottom. The red box indicates the first 49 amino acid residues of Ap5m1. Bottom: Detailed amino acid alignment showing high conservation in fish, amphibians and mammals. Note the high conservation of the Threonine (T) amino acid (red asterisk), which is mutated to Lysine (K) in ovyp35aluc (indicated in red in the lower panel, along with the splice site mutation, causing a premature stop codon). E. Structural superposition between zebrafish Ap5m1 (pink, amino acids 205–475) and the crystal structure of human Ap4m1 C-terminal domains (purple, amino acids 185–453). The RMSD value corresponds to 2.45 Å, thus demonstrating structural identity in overall 3D structure. |