Fig. 7
- ID
- ZDB-FIG-240708-19
- Publication
- Qin et al., 2024 - ABE-ultramax for high-efficiency biallelic adenine base editing in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
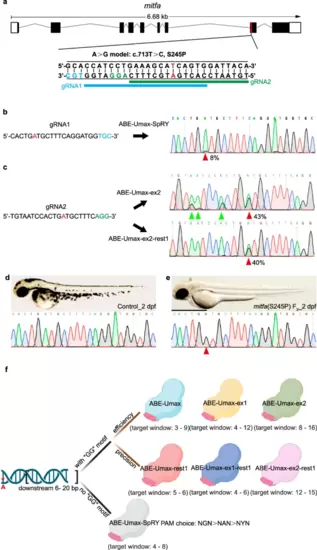
Disease modeling using ABE-Umax editors. a Generating A-to-G conversion in a disease-relevant zebrafish model (mitfa c.713 T > C). Protospacers are shown in black with PAM sequences in blue or green. The desired adenine for editing is highlighted in red. Potential gRNA target sites are marked with blue and green lines, respectively. b Sequencing results for mitfa (S245P) in ABE-Umax-SpRY-induced F0 generation. The desired adenine for editing is highlighted in red. Protospacers are shown in black with PAM sequences in blue. The red arrowhead indicates the expected nucleotide substitution. c Sequencing results for mitfa (S245P) in F0 generation induced by ABE-Umax-ex2 and ABE-Umax-ex2-rest1. The desired adenine for editing is highlighted in red. Protospacers are shown in black with PAM sequences in green. The red arrowhead indicates the expected nucleotide substitution. Bystander base substitutions are marked with green arrowheads in the Sanger sequencing chromatograms. d Lateral view of wild-type embryos at 2 dpf with sequencing results. Scale bar: 1 mm. e Lateral view of F2 homozygous embryos with the mitfa (S245P) mutation at 2 dpf, exhibiting pigmentation defects. The red arrowhead indicates the expected nucleotide substitution. Scale bar: 1 mm. f Selection of the right tool for A-to-G editing depends on the presence of a specific DNA sequence motif and can be selected as follows: Look for a GG motif within 6–20 base pairs downstream of the target adenine (red asterisks). Option 1: If a GG motif is present, (1) for high efficiency, consider ABE-Umax (window 3–9), ABE-Umax-ex1 (window 4–12), or ABE-Umax-ex2 (window 8–16); (2) for high precision, choose ABE-Umax-rest1 (window 5–6), ABE-Umax-ex1-rest1 (window 4–6), or ABE-Umax-ex2-rest1 (window 12–15). (3) If multiple tools meet your criteria, try them all to see which one works best. Option 2: If no GG motif is found, (1) use ABE-Umax-SpRY; (2) prioritize PAM sequences in this order: NGN > NAN > NYN; (3) position the edited adenine within the 4th–8th position of the editing window. The schematic diagrams in different colors represent different adenine-based editing tools. Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. |