Fig. 6
- ID
- ZDB-FIG-240708-18
- Publication
- Qin et al., 2024 - ABE-ultramax for high-efficiency biallelic adenine base editing in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
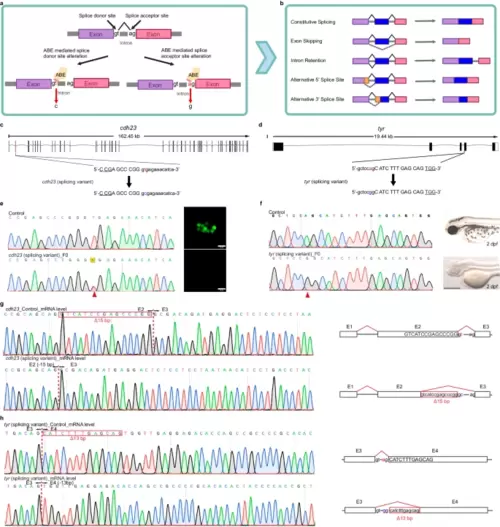
Editing of splice variants using ABE editors. a A schematic diagram of adenine base editing tools inducing splicing mutations. The colored bars (purple and pink) represent exons. Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. b Overview of possible common alternative splicing events. The mechanisms of the most common alternative splicing events, which include Exon Skipping, Intron Retention, Alternative 5′ splice site selection, and Alternative 3′ Splice Site selection, are shown. The colored bars (purple, blue, and pink) represent exons. Yellow bars represent the alternatively spliced exons, while the gray lines connecting them represent intronic segments. Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. c, d Schematic diagrams of the cdh23 (c) and tyr (d) splicing loci. The targeted sequence is displayed with the PAM underlined. The targeted nucleotides are highlighted in red, while the expected nucleotide changes are highlighted in blue. e Sequencing results and phenotype of the cdh23 splicing variant induced by ABE-Umax-ex2. The red arrowhead indicates the expected nucleotide substitutions. Neuromast hair cells labeled with Yo-Pro1 (green) are displayed adjacent to the chromatograms. Three independent experiments were repeated with similar results. Scale bars: 10 μm. f Sequencing results and phenotype of the tyr splicing variant induced by ABE-Umax. The red arrowhead indicates the expected nucleotide substitutions. Three independent experiments were repeated with similar results. Scale bars: 500 μm. g Sanger sequencing of cDNA demonstrates cdh23 splicing defects caused by ABE-Umax-ex2. The T-to-C substitution at the Exon 2 splicing donor site in cdh23 induces a 15 bp deletion at the mRNA level. The deletion sequence is highlighted in a red box. h Sanger sequencing reveals tyr splicing defects induced by ABE-Umax. The A-to-G substitution at the Exon 3 splicing acceptor site in tyr results in a 13 bp deletion at the mRNA level. The deletion sequence is highlighted in a red box. |