Fig. 2
- ID
- ZDB-FIG-240708-14
- Publication
- Qin et al., 2024 - ABE-ultramax for high-efficiency biallelic adenine base editing in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
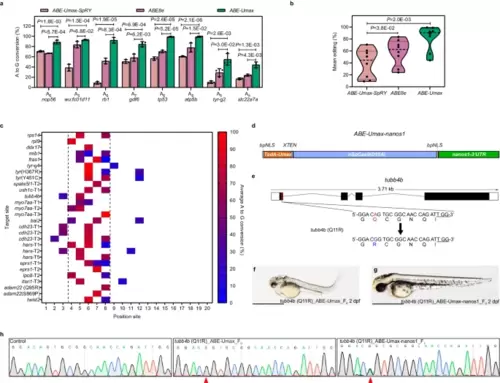
Efficient adenine base editing mediated by ABE-Umax. a The editing efficiency comparison among ABE-Umax-SpRY, ABE8e-SpRY, and ABE-Umax targeting eight different loci with NGG PAM. The editing base position within the gRNA is indicated by numbers. Values are presented as mean value ± standard deviation (SD), n = 3 biological replicates. Two-tailed paired t-test were performed (with P values marked). b Assessment of the mean editing efficiency of ABE-Umax-SpRY, ABE-Umax, and ABE8e Umax using the plot based on the data in Fig. 2a. Mean editing activity at each site is represented by individual data points, with the central dotted line indicating the overall mean. Two-tailed paired t-test were performed (with P values marked at the top of the violin plot.). c Heatmap depicting the average A-to-G editing efficiency of ABE-Umax across 28 target sites. Editing efficiency is represented by a color gradient, with red indicating 100% efficiency and blue indicating 0% efficiency. The black dotted line highlights the preferred editable range of ABE-Umax, between positions 4 and 8. d The mRNA construct of ABE-Umax-nanos1 for germline-specific adenine base editing. e Schematic diagram of the tubb4b locus. The targeted sequence is shown with the PAM underlined. The targeted nucleotide and amino acid are highlighted in red, while the expected changes in nucleotide and amino acid are highlighted in blue. f, g Phenotype of F0 embryos injected with tubb4b gRNA and ABE-Umax mRNA (f) or ABE-Umax-nanos1 mRNA (g). Lateral views of 2 dpf embryos are shown. Scale bar: 1 mm. h Sanger sequencing chromatograms confirm the A to G editing events in F0 injected embryos and F1. The desired mutation is indicated by a red arrowhead. All source data in this figure are provided as a source data file. |