Figure 2
- ID
- ZDB-FIG-230428-9
- Publication
- Frantz et al., 2023 - Pigment cell progenitor heterogeneity and reiteration of developmental signaling underlie melanocyte regeneration in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
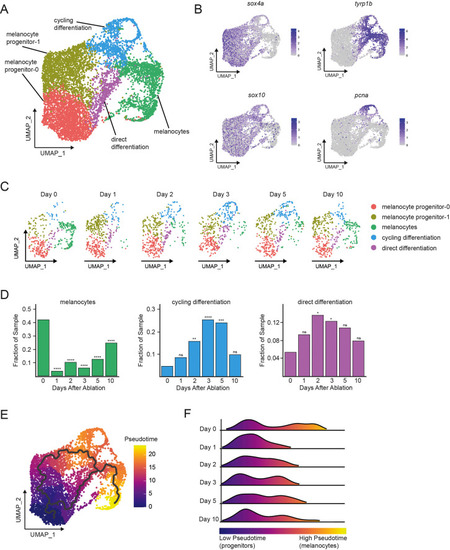
(A) Integrated subclustering of WT mitfa+aox5lo cells before, during, and after melanocyte regeneration (n = 5619 cells). Coloring is according to unsupervised clustering (Blondel et al., 2008; Stuart et al., 2019). Cell clusters are labeled based on gene expression patterns and inferred trajectories during regeneration, as revealed in panels (B–F) and Figure 2—figure supplement 1. Melanocyte and cycling differentiation clusters are the same as in Figure 1A, whereas the two precursor mitfa+aox5lo clusters from Figure 1A are now resolved into three clusters: melanocyte progenitor-0 (MP-0), melanocyte progenitor-1, and ‘direct differentiation’, the latter of which expresses the stem cell marker sox4a, the melanin biosynthesis gene tyrp1b and is pcna negative. (B) Expression of stem cell marker sox4a, melanin biosynthesis gene tyrp1b, pigment cell progenitor marker sox10, and cell cycle gene pcna as feature plots on the UMAP plot from (A). (C) Dynamic changes in subpopulations of WT mitfa+aox5lo cells. Biological sample runs were downsampled to a common number of total cells so shifts in cluster proportions could be readily visualized. (D) Quantification of proportion of cells per scRNAseq sample, comparing the indicated time point to day 0, in the melanocyte, cycling differentiation, and direct differentiation subpopulations during regeneration in WT animals (Day 0 = 7989, Day 1 = 4004, Day 2 = 4510, Day 3 = 5224, Day 5 = 3483, Day 10 = 4243 total cells per sample). p values calculated using differential proportion analysis (Farbehi et al., 2019), *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant. (E) Cellular trajectories, as determined by Monocle3 (Cao et al., 2019) projected onto the mitfa+aox5lo subcluster (left panel). Solid lines represent trajectories, with an origin in the MP-0 subpopulation and two distinct paths through different intermediate cell subpopulations. (F) These trajectories are then used to calculate pseudotime, which, as an approximation of biological time, reveals how low pseudotime progenitors (blue) progress across transitional cell types to high pseudotime melanocytes (yellow). Ridge plot of the distribution of pseudotime during regeneration. Height of ridge corresponds to number of cells at that pseudotime.
|