Figure 5
- ID
- ZDB-FIG-230220-5
- Publication
- Grepper et al., 2023 - Methodological advancements in organ-specific ectopic lipid quantitative characterization: Effects of high fat diet on muscle and liver intracellular lipids
- Other Figures
- All Figure Page
- Back to All Figure Page
|
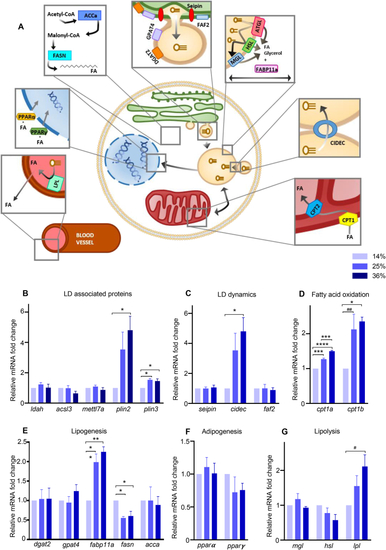
HFD altered lipid droplets associated pathways. (A) Schematic representation of target genes involved in LD coating proteins, dynamics, lipolysis, fatty acid oxidation, adipogenesis and lipogenesis. Lipoprotein lipase (lpl) hydrolyzes triglycerides and clears circulating lipids in the blood. Peroxisome proliferator-activated receptor α (pparα) and γ (pparγ) are transcription factors controlling lipid metabolism in various tissues. Acetyl-CoA carboxylase alpha (acca) catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, which is then synthetized into long chains FA by the fatty acid synthase (fasn). Glycerol-3-phosphate acyltransferase 4 (gpat4) relocalizes from the endoplasmic reticulum (ER) to forming LDs and catalyzes the first step of triacylglycerol (TAG) synthesis. Diglyceride acyltransferase 2 (dgat2) is responsible for the conversion of diacylglycerol (DAG) to TAG. Fas associated factor family member 2 (faf2) and seipin are ER proteins which form budding LDs. Adipose triglyceride lipase (atgl), hormone-sensitive lipase (hsl), and monoacylglycerol lipase (mgl) are lipases which breakdown triglycerides stored in LDs. Fatty acid binding protein 11a (fabp11a) is involved in lipid transport, storage and breakdown. Cell death Inducing DFFA like effector c (cidec) is involved in LD fusion and growth as well as apoptosis, while perilipin 2 and 3 (plin), acyl-CoA synthetase 3 (acsl3), lipid droplet associated hydrolase (ldah) and methyltransferase like 7A (mettl7a) are recruited to LD during maturation. Carnitine O-palmitoyltransferase 1a (cpt1a) and b (cpt1b) transform lipids to allow degradation through β-oxidation by mitochondria. (B-G) Gene expression measured by RT-qPCR. Data are normalized with the alpha subunit of the elongation factor-1 complex (ef1α). Bars represent mean ± SEM, n = 3 pools of 25 larvae from three independent experiments (One way ANOVA with Tukey HSD). For all panels: #p = 0.0513, ##p = 0.0529, ∗p < 0.05, ∗∗p ≤ 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. |