Fig. 6
- ID
- ZDB-FIG-221010-28
- Publication
- Chen et al., 2021 - A conserved residue in the P2X4 receptor has a nonconserved function in ATP recognition
- Other Figures
- All Figure Page
- Back to All Figure Page
|
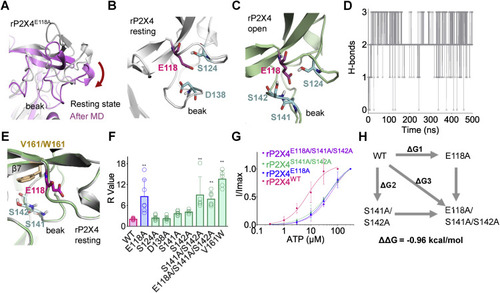
Figure 6. E118 interacts with S141/S142 in the beak region of rP2X4 receptors. A, downward motion of the head domain during 0.5-μs molecular dynamics (MD) simulation of rP2X4E118A in the resting state. B, polar amino acids of the head domain near E118 (within 5 Å) in rP2X4 in the resting (B) and open (C) states. D, the H-bond interactions between E118 and S141/S142 during 0.5-μs MD simulation of open structure of rP2X4. The trajectory was sampled every 200 ps. E, superimposed homology models of V161 and its tryptophan substitution (W161) to show the position of this residue (at rigid β7) near to S141, S142, and E118. F, summary of R values of WT and rP2X4 mutants. n = 15, 7, 8, 7, 7, 5, 5, 6, and 5, from left to right, respectively. One-way ANOVA with Bonferroni post-test (F(8, 56) = 19.98, p < 0.0001). The data on WT and E118A are the same as those displayed for Figure 5E. G, concentration–response curves of ATP in rP2X4WT, rP2X4E118A, rP2X4S141A/S142A, and rP2X4E118A/S141A/S142A. The solid lines are fitted by Hill equation (mean ± SEM, n = 3–5, EC50 = 51.5 ± 16.1 and 59.6 ± 13.2 μM, nH = 0.92 ± 0.14 and 1.07 ± 0.14 for S141A/S142A and S141A/S142A/E118A, respectively). The data of rP2X4WT and rP2X4E118A are the same as those displayed for Figure 4B and are shown here as controls. H, double-mutant cycle analysis between E118 and S141/S142. rP2X4, rat (Rattus norvegicus) P2X4. |