Fig. 1
- ID
- ZDB-FIG-221010-23
- Publication
- Chen et al., 2021 - A conserved residue in the P2X4 receptor has a nonconserved function in ATP recognition
- Other Figures
- All Figure Page
- Back to All Figure Page
|
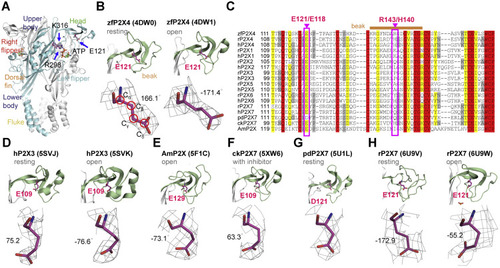
Figure 1. Side-chain orientations of E121 in zfP2X4 and equivalent residues in other P2X receptors. A, residues E121, R298, and K313 highlighted in the open structure of zfP2X4 (Protein data Bank [PBD] ID: 4DW1). The different domains are named head, dorsal fin (DF), left flipper (LF), right flipper (RF), body and fluke, respectively. B, zoom-in view of the location of E121 (upper) and 2Fo–Fc omit map for E121 (lower) in zfP2X4 at the resting (PDB ID: 4DW0, left) and open (right) states. Red circles indicate the atoms chosen to define the dihedral angle χCα–Cβ–Cγ–Cδ. C, the sequence alignment of the head domain in P2X family. Pink boxed regions highlight the highly conserved residue E121/E118 and nonconserved residue R143/H140 (zfP2X4/rP2X4 numbering) in P2X receptors. Sand bar indicates residues in the beak region. D–G, side-chain orientations of E109 in the apo (PDB ID: 5SVJ) and open (PDB ID: 5SVK) crystal structures of hP2X3, E129 in the open structure of AmP2X (PDB ID: 5F1C) (E), E109 in the crystal structure of ckP2X7 in complex with inhibitor (PDB ID: 5XW6) (F) and D121 in the apo crystal structure of pdP2X7 (PDB ID: 5U1L) (G). H, the side-chain orientation of E121 in the cryo-EM structure of rP2X7 at the resting (PDB ID: 6U9V) and open (PDB ID: 6U9W) states. rP2X4, rat (Rattus norvegicus) P2X4; zfP2X4, zebrafish (Danio rerio) P2X4. |