Figure 5—figure supplement 1—source data 1.
- ID
- ZDB-FIG-210220-34
- Publication
- Fontenas et al., 2021 - Spinal cord precursors utilize neural crest cell mechanisms to generate hybrid peripheral myelinating glia
- Other Figures
-
- Figure 1
- Figure 1—figure supplement 1—source data 1.
- Figure 1—figure supplement 2—source data 1.
- Figure 1—figure supplement 3.
- Figure 2—figure supplement 1.
- Figure 2—figure supplement 1.
- Figure 3—figure supplement 1.
- Figure 3—figure supplement 1.
- Figure 3—figure supplement 2.
- Figure 4
- Figure 5—figure supplement 1—source data 1.
- Figure 5—figure supplement 1—source data 1.
- Figure 6—figure supplement 1.
- Figure 6—figure supplement 1.
- Figure 6—figure supplement 2.
- Figure 7—figure supplement 1.
- Figure 7—figure supplement 1.
- Figure 8—figure supplement 1.
- Figure 8—figure supplement 1.
- Figure 8—figure supplement 2—source data 1.
- All Figure Page
- Back to All Figure Page
|
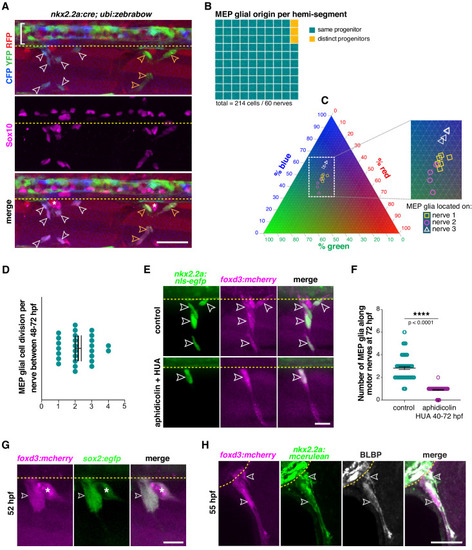
(A) Lateral view of Sox10 immunohistochemistry in a nkx2.2a:cre;ubi:zebrabow larvae showing Cre recombinase driven recombination (blue and green) in nkx2.2a+ cells at 4 dpf. Floor plate cells (bracket) and Sox10+ MEP glia along two distinct nerves (white and yellow outlined arrowheads respectively) labeled. (B) Percentage of MEP glia present along a single motor nerve sharing a common (96.67%) vs distinct neural tube precursor (3.33%); n = 214 MEP glia and n = 60 motor nerves in 20 animals. (C) Ternary plot showing the intensity profile of 14 MEP glia present on three adjacent motor nerves (white and yellow symbols in C correspond to white and yellow outlined arrowheads in A) for RFP, YFP and CFP, in percent. (D) Mean ± SEM of MEP glial cell divisions per motor nerve (2.19 ± 0.18) between 48 and 72 hpf; n = 27 nerves in 12 larvae. (E) DMSO control and aphidicolin/hydroxyurea (HUA)-treated nkx2.2a:nls-egfp;foxd3:mcherry larvae showing nkx2.2a+/ foxd3+ MEP glia (outlined arrowheads) along motor nerve root axons at 72 hpf. (F) Mean ± SEM of MEP glia along motor nerve roots at 72 hpf indicating 2.82 ± 0.12 MEP glia in control larvae (n = 79 hemi-segments from 10 larvae), and 0.90 ± 0.04 MEP glia in aphidicolin/HUA treated larvae (n = 89 hemi-segments from 10 larvae; p<0.0001). (G) Lateral view of a foxd3:mcherry;sox2:egfp larvae at 55 hpf showing foxd3+/sox2+ MEP glia (white outlined arrowhead) and DRG (asterisk). (H) Singe z plane transverse section of a BLBP immunostaining in foxd3:mcherry;nkx2.2a:mcerulean larvae showing BLBP+/foxd3+/nkx2.2a+ MEP glia (outlined arrowheads) outside the spinal cord along the motor nerve root at 55 hpf. Asterisks denote the DRG and yellow dashed lines denote the edge of the spinal cord. Scale bar, (A) 25 µm, (E, H) 20 µm, (G) 10 µm. |