Figure 3
- ID
- ZDB-FIG-200523-35
- Publication
- Koizumi et al., 2016 - The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction
- Other Figures
- All Figure Page
- Back to All Figure Page
|
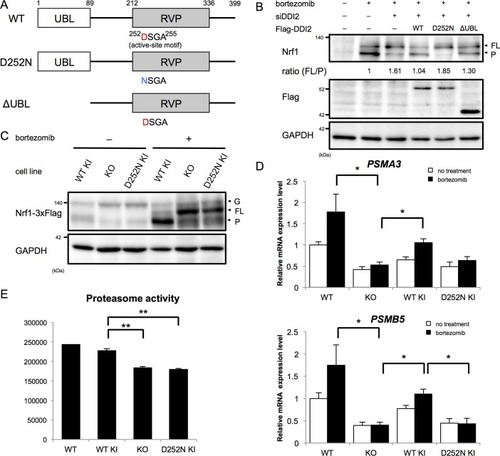
(A) Schematic diagram of wild-type (WT) and each mutant of DDI2. Ubiquitin-like (UBL) domain and retroviral protease-like (RVP) domain are represented as filled rectangles. The putative aspartyl protease active site amino acid sequence is shown. (B) HEK293A cells were transfected with DDI2 siRNA and after 24 hr were transfected with a plasmid encoding WT or mutant DDI2 shown in (A), followed by 50 nM bortezomib treatment for 14 hr before harvest. The signal intensity ratio of Nrf1 full-length form (FL) to the processed form (P) was calculated, where the ratio for bortezomib treatment alone was set as 1. (C) Immunoblotting of whole cell lysates of DDI2 WT knock-in (KI), DDI2 knockout (KO), and DDI2 D252N KI HCT116 cells. The cells transfected with Nrf1-3×Flag were treated with or without 50 nM borteaomib. (D) Relative mRNA expression of the proteasome genes PSMA3 and PSMB5 in WT, DDI2 KO, DDI2 WT KI, and DDI2 D252N KI HCT116 cells. mRNA levels of target genes were normalized by GUSB mRNA levels. The data represent mean + standard error of the mean (SEM) (n = 3, biological replicates). Statistical comparison was made by Tukey’s test (*p<0.05). (E) Proteasome peptidase activity of cell lysates of the indicated cell lines. The data represent mean + SEM (n = 3, biological replicates). Statistical comparison was made by Tukey’s test (**p<0.01). |