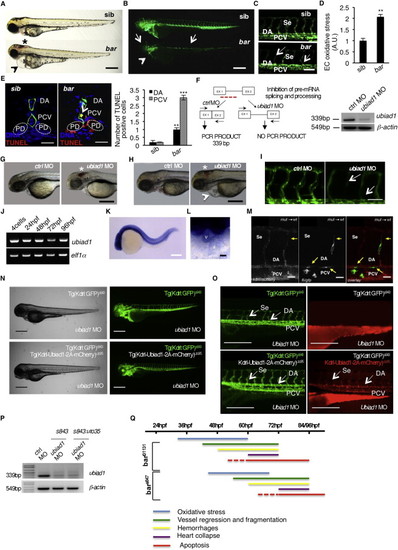
Barolo Alleles Phenotypes Are Characterized by Oxidative Stress Leading to Progressive Cardiovascular Failure, Related to Figure 1(A) Wild-type siblings s847 (sib) and barolos847 (bar) mutant embryos at 72 hpf. Vascular hemorrhages (asterisk) and heart failure (arrowhead) are visible in bars847 mutants. Scale bar, 300 �m.(B) Fluorescent micrographs of Tg(kdrl:GFP)s843 siblings (sib) and barolos847 (bar) mutants at 72 hpf. bars847 mutants show vascular integrity defects and regression both in brain and trunk vasculature (arrows). Endothelial cell regression is observed throughout the vascular network of bars847 mutants at 72 hpf. The collapsed endocardium is also evident (arrowhead). Scale bar, 300 �m.(C) Fluorescent micrographs representing magnification of trunk vasculature of Tg(kdrl:GFP)s843 sib s847 (sib) and barolos847 (bar) mutants at 72 hpf. In bars847 mutant�s endothelial cells are lost at the level of intersegmental vessels (Se), dorsal aorta (DA) and posterior cardinal vein (PCV) (arrows). Scale bar, 50 �m.(D) Histogram shows relative quantifications of oxidative stress in primary endothelial cells derived from Tg(kdrl:GFP)s843 sib s847 (sib) and bars847 (bar) mutant at 65 hpf. ROS levels in endothelial cells of zebrafish embryos were measured by flow cytometric analyses using the specific CellROX probe on Kdrl:GFP+ cells. ROS level is significantly higher in bars847 mutants than respective siblings. Results are shown as a mean of n = 3 independent experiments. Data are means � SD.(E) Staining of Tg(kdrl:GFP)s843 sibs847 (sib) and barolos847 (bar) mutant embryos at 80 hpf for TUNEL (red) and DNA (blue). Left: confocal transverse sections of trunk vasculature at the level of the 10th somite; right: histograms report numbers of dorsal aorta and posterior cardinal vein cells positive for TUNEL staining. Data are means � SD; p < 0.01, p < 0.001. TUNEL-positive cells were also present in pronephric ducts of bar mutants. Scale bars, 100 �m.(F) Validation of in vivo ubiad1 knockdown by morpholino injection. Left: schematic diagram shows the molecular mechanism of ubiad1splice-morpholino (ubiad1 MO; dashed line). ubiad1 MO targeting the splicing site between exon1 (EX1) and intron1 blocks ubiad1 pre-mRNA maturation and leads to ubiad1 non-coding mRNA. Molecular mechanism of control non-targeting morpholino (ctrl MO) is also indicated. Primers used for RT-PCR validation are localized on exons 1 and 2 (EX1-EX2; black arrows). Right: RT-PCR analysis was performed on cDNA from zebrafish embryos 72 hr post microinjection. Amplification of 339 bp is possible only for cDNA from ctrl-injected embryos, whereas no amplification is expected from ubiad1MO-injected embryos. Compared to ctrl MO, ubiad1 MO show reduced 339 bp PCR products.Representative bright-field images of ubiad1 morphants (ubiad1 MO) and relative control (ctrl MO) at 48 hpf (G) and 72 hpf (H). ubiad1 morphants such as bar mutants are characterized by brain hemorrhages (asterisk) and stringy heart morphology (arrowhead). Scale bar, 100 �m.(I) Fluorescent micrographs of trunk vasculature (between 14th and 18th somites) of Tg (kdrl:GFP)s843ubiad1 morphants (ubiad1 MO) and control (ctrl MO) injected embryos at 72 hpf. Knockdown of ubiad1 led to loss of intersegmental vessels integrity (arrows) as observed in bar mutants. Scale bar, 75 �m.(J) RT-PCR analyses for ubiad1 mRNA expression and ef1� (as control) on cDNA from wild-type embryos at different developmental stages. Samples are from 4-cell stage to 96 hpf stage.(K) Whole-mount ISH analyses of wild-type embryo at 24 hpf show ubiad1 mRNA expression throughout the embryo. Scale bar, 250 �m.(L) Magnification on heart of whole-mount ISH analyses of wild-type embryo at 48 hpf. Specific expression of ubiad1 mRNA can be detected in the developing heart. A, atrium; V, ventricle. Scale bar, 50 �m.(M) Cell transplantation experiments in barolo mutant embryos. Confocal fluorescent image shows chimeric vessels made of endothelial cells from Tg(fli:GFP)y1bart31131 mutant embryos (donor) that have been transplanted at pre-dome stage into Tg(kdrl: mCherry)uto2 wild-type (wt) embryos (acceptor) and visualized at 48 hpf. Transplanted mutant endothelial cells (green fluorescence) show retracting and collapsed morphology (arrows), also at the level of intersegmental vessel (Se) and dorsal aorta (DA) suggesting ubiad1 acts cell autonomously. Left: mCherry channel corresponding to Tg(kdrl: mCherry)uto2 wild-type cells. Middle: GFP channel corresponding to Tg(fli:GFP)y1bart31131 mutant endothelial cells. Right: Merge image of mCherry and GFP channels. Se, intersomitic vessels; DA, dorsal aorta; PCV, posterior cardinal vein. Scale bar, 20 �m.(N) Bright field (left) and corresponding fluorescent micrographs (right) of Tg(kdrl:GFP)s843 and Tg [(kdrl-Ubiad1-2A-mCherry)uto35; (kdrl:GFP)s843 ] wild-type embryos injected with ubiad1 splice-morpholino (ubiad1 MO). After morpholino injection Tg(kdrl:GFP)s843 embryos show characteristic barolo phenotype (upper). On the contrary, after ubiad1 morpholino injection Tg [(kdrl-Ubiad1-2A-mCherry)uto35; (kdrl:GFP)s843 ] show wild-type phenotype (lower). Scale bar, 400 �m.(O) Magnification of trunk vasculature of embryos showed in (N). Green fluorescence shows endothelial cells. Red fluorescence, which is exclusive of uto35 line, indicates expression of exogenous Ubiad1 in endothelial cells (lower right). Injection of ubiad1 morpholino in Tg(kdrl:GFP)s843 induce bar phenotype characterized by intersegmental vessels (Se) disintegration and collapsed dorsal aorta (DA) (upper left; arrows). Injection of ubiad1 morpholino in Tg [(kdrl-Ubiad1-2A-mCherry)uto35;(kdrl:GFP)s843 ] do not exert any vascular phenotype (lower left; arrowheads) indicating that endothelial-specific expression of Ubiad1 is sufficient to prevent barolo cardiovascular phenotype. All together these data support that Ubiad1 acts cell autonomously in vascular cells. Scale bar, 150 �m.(P) Validation of ubiad1 knockdown by morpholino injection, related to embryos in (N and O). RT-PCR analysis was performed on cDNA from zebrafish embryos at 72 hpf. Amplification of 339bp is possible only for cDNA from ctrl MO-injected embryos (ctrl MO), whereas no amplification is expected from ubiad1MO-injected embryos (ubiad1 MO). Compared to ctrl MO, ubiad1 MO show reduced 339bp PCR products in Tg(kdrl:GFP)s843 (s843) as well as in Tg [(kdrl-Ubiad1-2A-mCherry)uto35; (kdrl:GFP)s843 ] (s843:uto35) morphants. RT-PCR strategy is showed in (F).(Q) Chronological documentation of barolo phenotypes onset. bart31131 mutant embryos phenotype starts with an increase of oxidative stress (detectable from 32 hpf until 60 hpf) that causes a progressive cardiovascular breakdown characterized by vessels regression and fragmentation (starting from 40 hpf), hemorrhages (from 48 hpf) and heart failure (from 60 hpf). barolo cardiovascular degeneration ends up with heart and vascular cells apoptosis that become evident at molecular levels starting from 60 hpf. Definitive signs of apoptosis can be scored at 72 hpf.bars847 shows the same progressive cardiovascular failure features of as bart3113, but delayed of 12 hr in the same experimental conditions. Both barolo alleles are not able to survive over 96 hpf.
|

