Fig. 2
- ID
- ZDB-FIG-120126-61
- Publication
- Lindeman et al., 2011 - Prepatterning of Developmental Gene Expression by Modified Histones before Zygotic Genome Activation
- Other Figures
- All Figure Page
- Back to All Figure Page
|
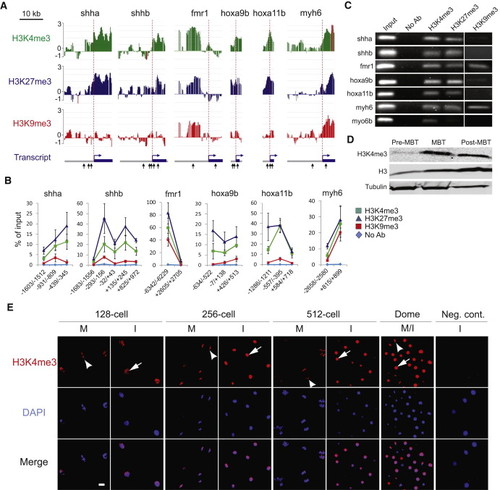
Enrichment in H3K4me3, H3K27me3, and H3K9me3 at the Pre-MBT Stage(A) H3K4me3, H3K27me3, and H3K9me3 ChIP-chip profiles on indicated genes at the pre-MBT stage (MA2C scores). Genes were ranked as follows, in decreasing order, among the 1081 genes enriched in H3K4me3 pre-MBT: shha (107), fmr1 (226), hoxa9b (284), hoxa11b (464), myh6 (679); shhb was also selected for its H3K4me3 marking (though not enrichment) in the absence of H3K9me3. Arrows (bottom track) mark amplicons examined by ChIP-qPCR in (B).(B) ChIP-qPCR validation of H3K4me3, H3K27me3, and H3K9me3 pre-MBT marking on indicated genes (mean ± SD of three ChIPs with each antibody).(C) Native ChIP-PCR validation of H3K4me3, H3K27me3, and H3K9me3 occupancy.(D) Western blot analysis of H3K4me3 in developing embryos.(E) Immunofluorescence detection of H3K4me3 in the animal pole of embryos at indicated stages. Arrows and arrowheads point to labeled nuclei and metaphases, respectively. Signal intensity is adjusted to 128-cell images. Time points: 128-cell, 2.2 hpf; 256-cell, 2.5 hpf; 512-cell, 2.7 hpf; Dome, 4.3 hpf. I, interphase; M, metaphase. Scale bar, 10 μm. |
| Genes: | |
|---|---|
| Fish: | |
| Anatomical Term: | |
| Stage: | 256-cell |
Reprinted from Developmental Cell, 21(6), Lindeman, L.C., Andersen, I.S., Reiner, A.H., Li, N., Aanes, H., Ostrup, O., Winata, C., Mathavan, S., Muller, F., Aleström, P., and Collas, P., Prepatterning of Developmental Gene Expression by Modified Histones before Zygotic Genome Activation, 993-1004, Copyright (2011) with permission from Elsevier. Full text @ Dev. Cell