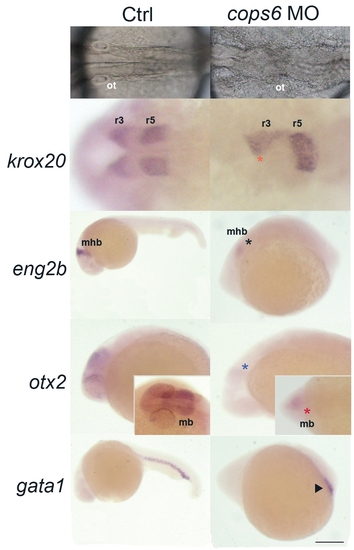
Zebrafish cops6 plays a role in brain development, but not in blood vessel formation. (A,B) cops6 morphants in an AB wild-type background showed a brain defect at the prim-5 stage. The hindbrain was not well formed in (B) cops6 morphants when compared to (A) control; dorsal view. (C,D) Expression of krox20, a marker of rhombomeres three and five; dorsal view. Reduced size in rhombomere three (orange asterisk) and fused rhombomere five expression due to unfolded hindbrain were found. (E,F) eng2b expression that indicates the mid-hindbrain boundary (mhb) was reduced in cops6 morphants (black asterisk). (F) cops6 morphants had a smaller mhb when compared to (E) control; lateral view. (G,H) otx2 expression was reduced in cops6 morphants (blue asterisk in lateral view; red asterisk in dorsal view). The size of the midbrain was decreased in (H) cops6 morphants in comparison to (G) control. (I,J) Presumptive blood marker (gata1) did not show any significant differences between (I) control and (J) cops6 morphants. They both form a normal blood island (triangle mark), which suggests that cops6 is not required for blood development. mb: midbrain; mhb: mid-hindbrain boundary; ot: otic vesicle; r3/r5: rhombomere three/five. All photos are with head to the left. Scale bar: 48 μm (A,B), 100 μm (C,D), 265 μm (E,I), 150 μm (F,J) and 170 μm (G,H).
|

