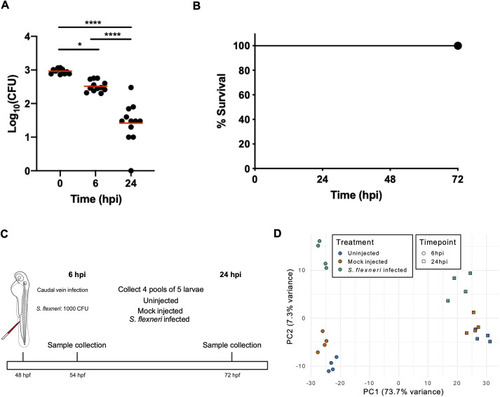
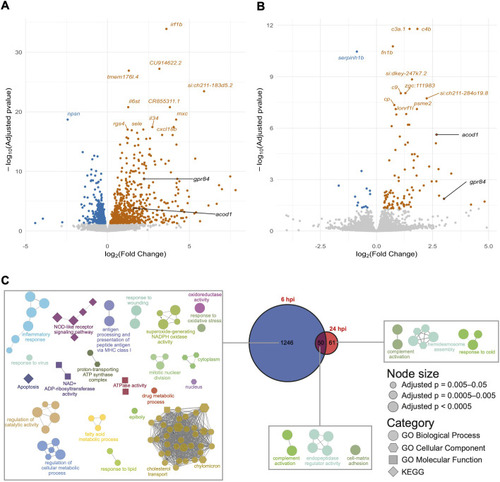
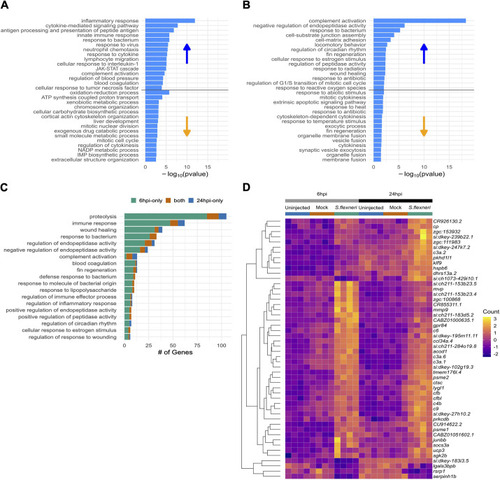
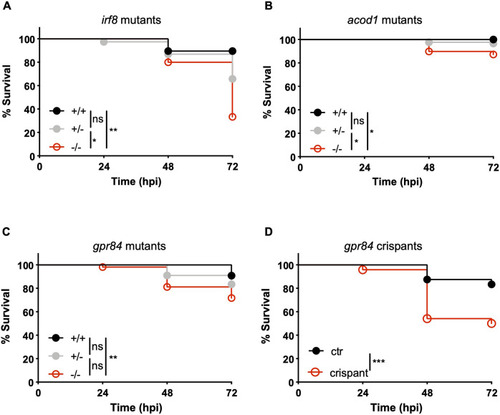
- Title
-
Transcriptional profiling of zebrafish identifies host factors controlling susceptibility to Shigella flexneri
- Authors
- Torraca, V., White, R.J., Sealy, I.M., Mazon-Moya, M., Duggan, G., Willis, A., Busch-Nentwich, E.M., Mostowy, S.
- Source
- Full text @ Dis. Model. Mech.
|
|
|
|
|
|
|
|