- Title
-
Infection-experienced HSPCs protect against infections by generating neutrophils with enhanced mitochondrial bactericidal activity
- Authors
- Darroch, H., Keerthisinghe, P., Sung, Y.J., Rolland, L., Prankerd-Gough, A., Crosier, P.S., Astin, J.W., Hall, C.J.
- Source
- Full text @ Sci Adv
|
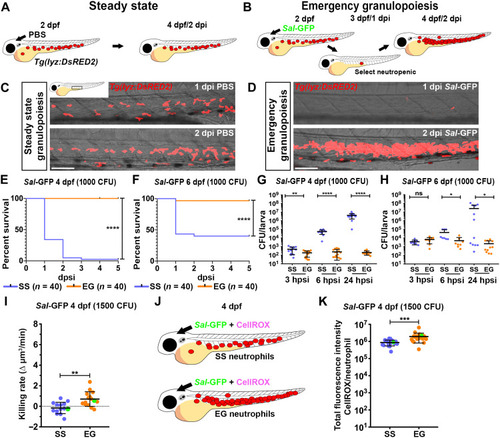
EG larvae have elevated survival to subsequent bacterial challenge and have neutrophils with enhanced bactericidal activity. ( |
|
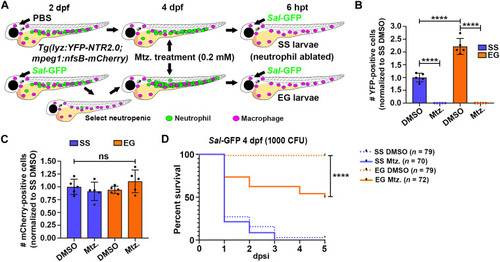
Ablating EG neutrophils reduces survival to subsequent infection. ( |
|
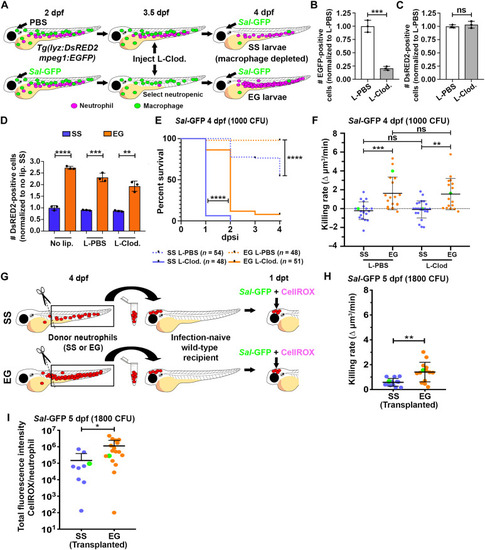
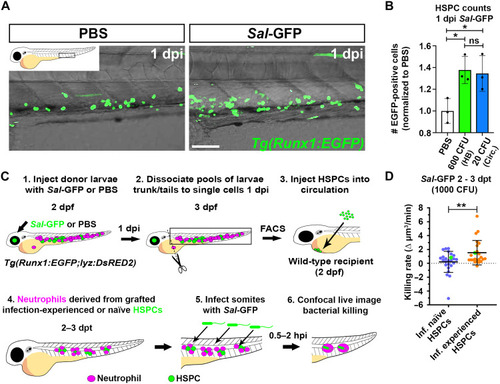
EG neutrophils maintain elevated killing rates in macrophage-depleted larvae and when transplanted into infection-naïve recipients. ( |
|
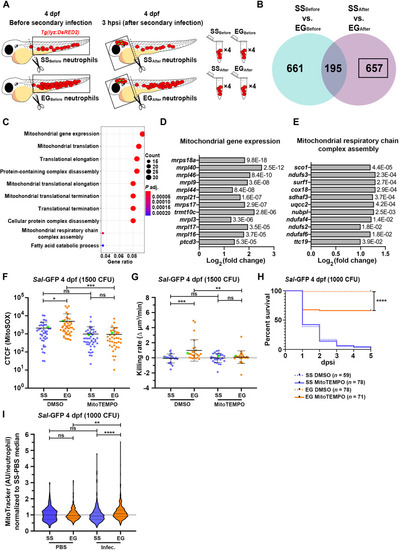
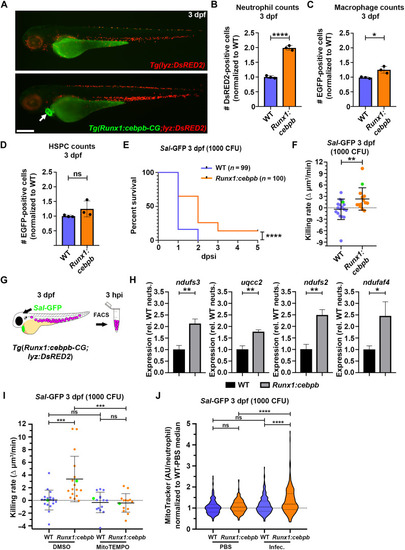
EG neutrophils enhance expression of mitochondria-associated genes following infection, use mtROS for their enhanced bactericidal activity and have greater mitochondrial mass. ( |
|
Infection-experienced HSPCs generate neutrophils with elevated bactericidal activity. ( |
|
Larvae with ( |